Benchmarks for cluster¶
KMeans-minimadelon¶
Benchmark setup
from sklearn.cluster import KMeans from deps import load_data kwargs = {'n_clusters': 9} X, y, X_t, y_t = load_data('minimadelon') obj = KMeans(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
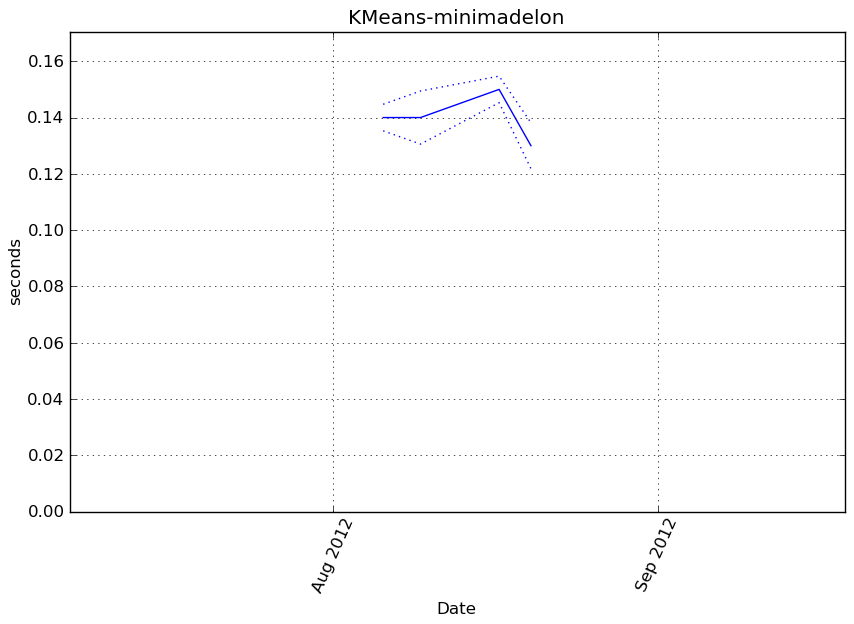
Memory usage
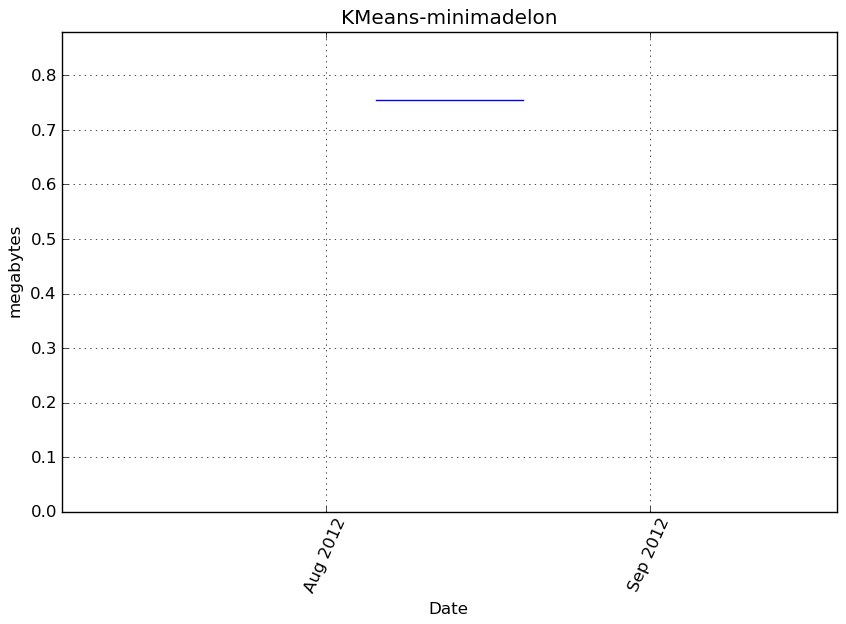
Additional output
cProfile
16149 function calls in 0.164 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.164 0.164 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.164 0.164 <f>:1(<module>)
1 0.000 0.000 0.164 0.164 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:738(fit)
1 0.000 0.000 0.164 0.164 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:151(k_means)
10 0.002 0.000 0.162 0.016 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:303(_kmeans_single)
117 0.011 0.000 0.091 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:101(euclidean_distances)
10 0.000 0.000 0.091 0.009 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:525(_init_centroids)
10 0.017 0.002 0.090 0.009 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:36(_k_init)
27 0.027 0.001 0.039 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:478(_centers)
117 0.001 0.000 0.035 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
117 0.002 0.000 0.032 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:70(safe_sparse_dot)
27 0.001 0.000 0.029 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:431(_labels_inertia)
352 0.001 0.000 0.029 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
27 0.004 0.000 0.028 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:414(_labels_inertia_precompute_dense)
117 0.028 0.000 0.028 0.000 {numpy.core._dotblas.dot}
586 0.006 0.000 0.021 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
1088 0.020 0.000 0.020 0.000 {method 'sum' of 'numpy.ndarray' objects}
1448 0.002 0.000 0.018 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
1448 0.010 0.000 0.015 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
234 0.001 0.000 0.014 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
352 0.001 0.000 0.011 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
245 0.010 0.000 0.010 0.000 {method 'mean' of 'numpy.ndarray' objects}
352 0.003 0.000 0.005 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
144 0.000 0.000 0.003 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
2896 0.003 0.000 0.003 0.000 {method 'split' of 'str' objects}
586 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
938 0.002 0.000 0.002 0.000 {numpy.core.multiarray.array}
243 0.000 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
1656 0.002 0.000 0.002 0.000 {isinstance}
243 0.002 0.000 0.002 0.000 {method 'any' of 'numpy.ndarray' objects}
352 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
80 0.001 0.000 0.001 0.000 {method 'random_sample' of 'mtrand.RandomState' objects}
118 0.001 0.000 0.001 0.000 {numpy.core.multiarray.empty}
89 0.001 0.000 0.001 0.000 {method 'copy' of 'numpy.ndarray' objects}
1 0.000 0.000 0.001 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:142(_tolerance)
80 0.001 0.000 0.001 0.000 {method 'cumsum' of 'numpy.ndarray' objects}
80 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:757(searchsorted)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2470(var)
1 0.000 0.000 0.000 0.000 {method 'var' of 'numpy.ndarray' objects}
1290 0.000 0.000 0.000 0.000 {len}
27 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:710(_check_fit_data)
80 0.000 0.000 0.000 0.000 {method 'searchsorted' of 'numpy.ndarray' objects}
81 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
352 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
32 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
65 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:404(_squared_norms)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:33(as_float_array)
10 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2299(mean)
10 0.000 0.000 0.000 0.000 {method 'randint' of 'mtrand.RandomState' objects}
1 0.000 0.000 0.000 0.000 {hasattr}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/k_means_.py
Function: fit at line 738
Total time: 0.190057 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
738 def fit(self, X, y=None):
739 """Compute k-means"""
740
741 1 4 4.0 0.0 if not self.k is None:
742 n_clusters = self.k
743 warnings.warn("Parameter k was renamed to n_clusters",
744 DeprecationWarning, stacklevel=2)
745 self.n_clusters = n_clusters
746 else:
747 1 3 3.0 0.0 n_clusters = self.n_clusters
748
749 1 9 9.0 0.0 self.random_state = check_random_state(self.random_state)
750 1 147 147.0 0.1 X = self._check_fit_data(X)
751
752 1 3 3.0 0.0 self.cluster_centers_, self.labels_, self.inertia_ = k_means(
753 1 3 3.0 0.0 X, n_clusters=n_clusters, init=self.init, n_init=self.n_init,
754 1 2 2.0 0.0 max_iter=self.max_iter, verbose=self.verbose,
755 1 2 2.0 0.0 precompute_distances=self.precompute_distances,
756 1 3 3.0 0.0 tol=self.tol, random_state=self.random_state, copy_x=self.copy_x,
757 1 189876 189876.0 99.9 n_jobs=self.n_jobs)
758 1 5 5.0 0.0 return self
KMeans-blobs¶
Benchmark setup
from sklearn.cluster import KMeans from deps import load_data kwargs = {'n_clusters': 9} X, y, X_t, y_t = load_data('blobs') obj = KMeans(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
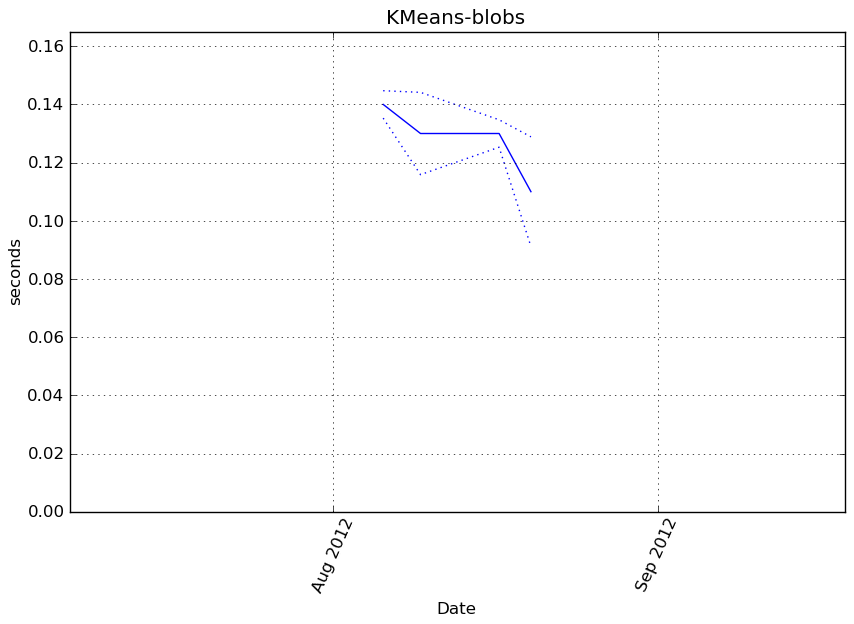
Memory usage
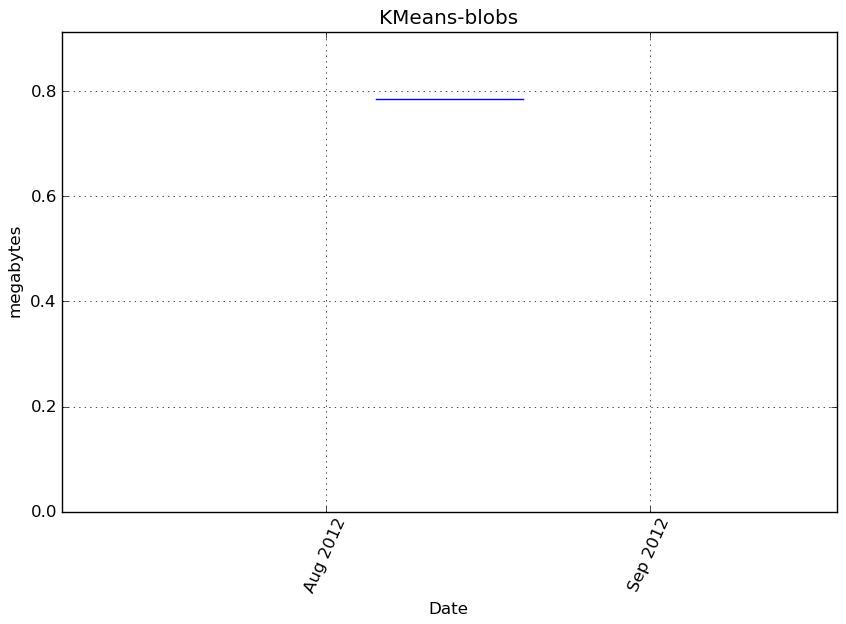
Additional output
cProfile
15637 function calls in 0.093 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.093 0.093 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.093 0.093 <f>:1(<module>)
1 0.000 0.000 0.093 0.093 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:738(fit)
1 0.000 0.000 0.093 0.093 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:151(k_means)
10 0.001 0.000 0.092 0.009 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:303(_kmeans_single)
114 0.008 0.000 0.054 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:101(euclidean_distances)
10 0.000 0.000 0.053 0.005 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:525(_init_centroids)
10 0.007 0.001 0.053 0.005 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:36(_k_init)
24 0.016 0.001 0.021 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:478(_centers)
114 0.001 0.000 0.021 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
114 0.001 0.000 0.018 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:70(safe_sparse_dot)
24 0.000 0.000 0.018 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:431(_labels_inertia)
343 0.001 0.000 0.018 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
24 0.004 0.000 0.017 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:414(_labels_inertia_precompute_dense)
114 0.016 0.000 0.016 0.000 {numpy.core._dotblas.dot}
571 0.004 0.000 0.012 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
1064 0.012 0.000 0.012 0.000 {method 'sum' of 'numpy.ndarray' objects}
1409 0.001 0.000 0.011 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
1409 0.006 0.000 0.009 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
228 0.001 0.000 0.008 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
343 0.001 0.000 0.007 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
218 0.004 0.000 0.004 0.000 {method 'mean' of 'numpy.ndarray' objects}
343 0.002 0.000 0.003 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
2818 0.002 0.000 0.002 0.000 {method 'split' of 'str' objects}
138 0.000 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
571 0.001 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
914 0.001 0.000 0.001 0.000 {numpy.core.multiarray.array}
1611 0.001 0.000 0.001 0.000 {isinstance}
216 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
343 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
216 0.001 0.000 0.001 0.000 {method 'any' of 'numpy.ndarray' objects}
80 0.001 0.000 0.001 0.000 {method 'cumsum' of 'numpy.ndarray' objects}
80 0.000 0.000 0.000 0.000 {method 'random_sample' of 'mtrand.RandomState' objects}
106 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
80 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:757(searchsorted)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:142(_tolerance)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2470(var)
1 0.000 0.000 0.000 0.000 {method 'var' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:710(_check_fit_data)
80 0.000 0.000 0.000 0.000 {method 'searchsorted' of 'numpy.ndarray' objects}
1257 0.000 0.000 0.000 0.000 {len}
24 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
78 0.000 0.000 0.000 0.000 {method 'copy' of 'numpy.ndarray' objects}
343 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
72 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
32 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:404(_squared_norms)
59 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:33(as_float_array)
10 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2299(mean)
10 0.000 0.000 0.000 0.000 {method 'randint' of 'mtrand.RandomState' objects}
1 0.000 0.000 0.000 0.000 {hasattr}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/k_means_.py
Function: fit at line 738
Total time: 0.107761 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
738 def fit(self, X, y=None):
739 """Compute k-means"""
740
741 1 2 2.0 0.0 if not self.k is None:
742 n_clusters = self.k
743 warnings.warn("Parameter k was renamed to n_clusters",
744 DeprecationWarning, stacklevel=2)
745 self.n_clusters = n_clusters
746 else:
747 1 2 2.0 0.0 n_clusters = self.n_clusters
748
749 1 6 6.0 0.0 self.random_state = check_random_state(self.random_state)
750 1 97 97.0 0.1 X = self._check_fit_data(X)
751
752 1 1 1.0 0.0 self.cluster_centers_, self.labels_, self.inertia_ = k_means(
753 1 2 2.0 0.0 X, n_clusters=n_clusters, init=self.init, n_init=self.n_init,
754 1 2 2.0 0.0 max_iter=self.max_iter, verbose=self.verbose,
755 1 2 2.0 0.0 precompute_distances=self.precompute_distances,
756 1 2 2.0 0.0 tol=self.tol, random_state=self.random_state, copy_x=self.copy_x,
757 1 107643 107643.0 99.9 n_jobs=self.n_jobs)
758 1 2 2.0 0.0 return self
MiniBatchKMeans-minimadelon¶
Benchmark setup
from sklearn.cluster import MiniBatchKMeans from deps import load_data kwargs = {'n_clusters': 9} X, y, X_t, y_t = load_data('minimadelon') obj = MiniBatchKMeans(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
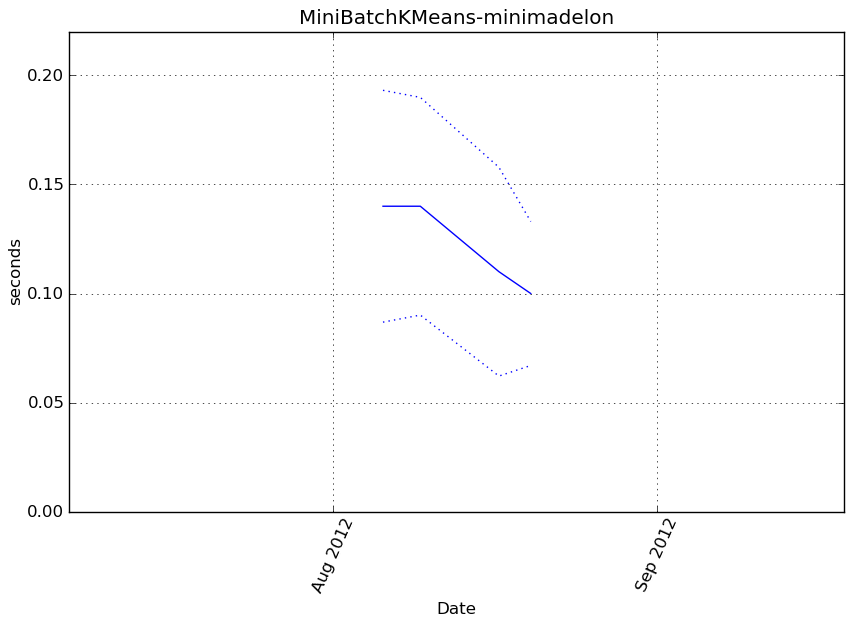
Memory usage
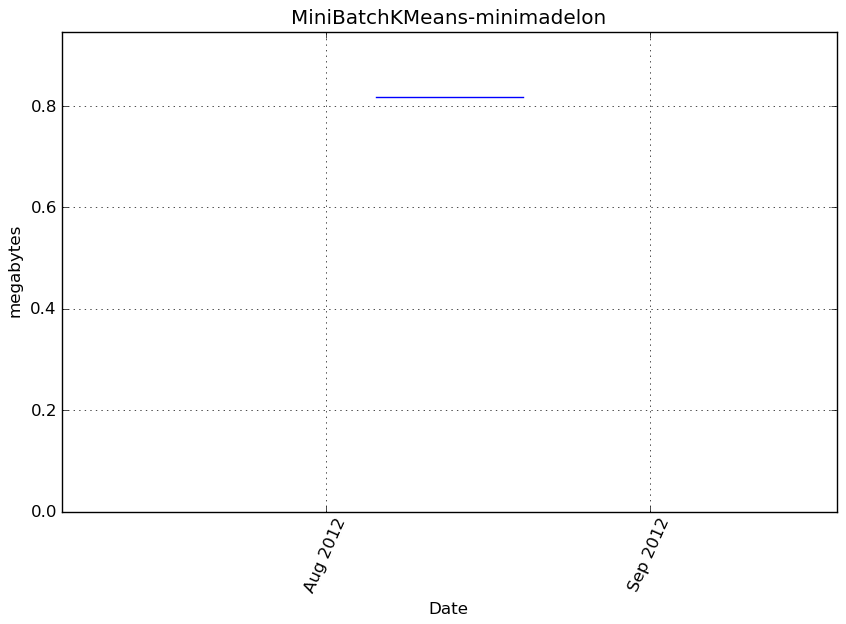
Additional output
cProfile
10159 function calls in 0.302 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.302 0.302 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.302 0.302 <f>:1(<module>)
1 0.108 0.108 0.302 0.302 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:1067(fit)
35 0.125 0.004 0.172 0.005 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:830(_mini_batch_step)
66 0.004 0.000 0.046 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:101(euclidean_distances)
39 0.001 0.000 0.039 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:431(_labels_inertia)
39 0.004 0.000 0.037 0.001 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:414(_labels_inertia_precompute_dense)
1152 0.020 0.000 0.020 0.000 {method 'sum' of 'numpy.ndarray' objects}
66 0.001 0.000 0.020 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:70(safe_sparse_dot)
66 0.018 0.000 0.018 0.000 {numpy.core._dotblas.dot}
3 0.000 0.000 0.018 0.006 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:525(_init_centroids)
3 0.003 0.001 0.018 0.006 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:36(_k_init)
66 0.001 0.000 0.016 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
198 0.001 0.000 0.012 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
330 0.002 0.000 0.011 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
368 0.001 0.000 0.010 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
132 0.000 0.000 0.007 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
832 0.001 0.000 0.006 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
832 0.004 0.000 0.006 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
198 0.001 0.000 0.004 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
198 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
1212 0.001 0.000 0.001 0.000 {isinstance}
1664 0.001 0.000 0.001 0.000 {method 'split' of 'str' objects}
330 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
529 0.001 0.000 0.001 0.000 {numpy.core.multiarray.array}
198 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
32 0.001 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:898(_mini_batch_convergence)
120 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
39 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
117 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
33 0.000 0.000 0.000 0.000 {method 'random_integers' of 'mtrand.RandomState' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:404(_squared_norms)
727 0.000 0.000 0.000 0.000 {len}
24 0.000 0.000 0.000 0.000 {method 'random_sample' of 'mtrand.RandomState' objects}
24 0.000 0.000 0.000 0.000 {method 'cumsum' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:94(check_arrays)
199 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
24 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:757(searchsorted)
75 0.000 0.000 0.000 0.000 {range}
24 0.000 0.000 0.000 0.000 {method 'searchsorted' of 'numpy.ndarray' objects}
7 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
9 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
128 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:289(ascontiguousarray)
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:87(_num_samples)
5 0.000 0.000 0.000 0.000 {hasattr}
3 0.000 0.000 0.000 0.000 {method 'randint' of 'mtrand.RandomState' objects}
4 0.000 0.000 0.000 0.000 {method 'pop' of 'dict' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/k_means_.py
Function: fit at line 1067
Total time: 0.513636 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
1067 def fit(self, X, y=None):
1068 """Compute the centroids on X by chunking it into mini-batches.
1069
1070 Parameters
1071 ----------
1072 X: array-like, shape = [n_samples, n_features]
1073 Coordinates of the data points to cluster
1074 """
1075 1 8 8.0 0.0 self.random_state = check_random_state(self.random_state)
1076 1 3 3.0 0.0 X = check_arrays(X, sparse_format="csr", copy=False,
1077 1 57 57.0 0.0 check_ccontiguous=True, dtype=np.float64)[0]
1078 1 2 2.0 0.0 n_samples, n_features = X.shape
1079 1 2 2.0 0.0 if n_samples < self.n_clusters:
1080 raise ValueError("Number of samples smaller than number "\
1081 "of clusters.")
1082
1083 1 5 5.0 0.0 if hasattr(self.init, '__array__'):
1084 self.init = np.ascontiguousarray(self.init, dtype=np.float64)
1085
1086 1 84 84.0 0.0 x_squared_norms = _squared_norms(X)
1087
1088 1 3 3.0 0.0 if self.tol > 0.0:
1089 tol = _tolerance(X, self.tol)
1090
1091 # using tol-based early stopping needs the allocation of a
1092 # dedicated before which can be expensive for high dim data:
1093 # hence we allocate it outside of the main loop
1094 old_center_buffer = np.zeros(n_features, np.double)
1095 else:
1096 1 2 2.0 0.0 tol = 0.0
1097 # no need for the center buffer if tol-based early stopping is
1098 # disabled
1099 1 8 8.0 0.0 old_center_buffer = np.zeros(0, np.double)
1100
1101 1 6 6.0 0.0 distances = np.zeros(self.batch_size, dtype=np.float64)
1102 1 13 13.0 0.0 n_batches = int(np.ceil(float(n_samples) / self.batch_size))
1103 1 3 3.0 0.0 n_iterations = int(self.max_iter * n_batches)
1104
1105 1 2 2.0 0.0 init_size = self.init_size
1106 1 2 2.0 0.0 if init_size is None:
1107 1 2 2.0 0.0 init_size = 3 * self.batch_size
1108 1 2 2.0 0.0 if init_size > n_samples:
1109 1 2 2.0 0.0 init_size = n_samples
1110 1 2 2.0 0.0 self.init_size_ = init_size
1111
1112 1 2 2.0 0.0 validation_indices = self.random_state.random_integers(
1113 1 8 8.0 0.0 0, n_samples - 1, init_size)
1114 1 512 512.0 0.1 X_valid = X[validation_indices]
1115 1 7 7.0 0.0 x_squared_norms_valid = x_squared_norms[validation_indices]
1116
1117 # perform several inits with random sub-sets
1118 1 2 2.0 0.0 best_inertia = None
1119 4 12 3.0 0.0 for init_idx in range(self.n_init):
1120 3 6 2.0 0.0 if self.verbose:
1121 print "Init %d/%d with method: %s" % (
1122 init_idx + 1, self.n_init, self.init)
1123 3 22 7.3 0.0 counts = np.zeros(self.n_clusters, dtype=np.int32)
1124
1125 # TODO: once the `k_means` function works with sparse input we
1126 # should refactor the following init to use it instead.
1127
1128 # Initialize the centers using only a fraction of the data as we
1129 # expect n_samples to be very large when using MiniBatchKMeans
1130 3 6 2.0 0.0 cluster_centers = _init_centroids(
1131 3 6 2.0 0.0 X, self.n_clusters, self.init,
1132 3 6 2.0 0.0 random_state=self.random_state,
1133 3 6 2.0 0.0 x_squared_norms=x_squared_norms,
1134 3 78703 26234.3 15.3 init_size=init_size)
1135
1136 # Compute the label assignement on the init dataset
1137 3 10 3.3 0.0 batch_inertia, centers_squared_diff = _mini_batch_step(
1138 3 24 8.0 0.0 X_valid, x_squared_norms[validation_indices],
1139 3 8 2.7 0.0 cluster_centers, counts, old_center_buffer, False,
1140 3 5827 1942.3 1.1 distances=distances)
1141
1142 # Keep only the best cluster centers across independent inits on
1143 # the common validation set
1144 3 11 3.7 0.0 _, inertia = _labels_inertia(X_valid, x_squared_norms_valid,
1145 3 2410 803.3 0.5 cluster_centers)
1146 3 11 3.7 0.0 if self.verbose:
1147 print "Inertia for init %d/%d: %f" % (
1148 init_idx + 1, self.n_init, inertia)
1149 3 9 3.0 0.0 if best_inertia is None or inertia < best_inertia:
1150 1 2 2.0 0.0 self.cluster_centers_ = cluster_centers
1151 1 2 2.0 0.0 self.counts_ = counts
1152 1 2 2.0 0.0 best_inertia = inertia
1153
1154 # Empty context to be used inplace by the convergence check routine
1155 1 2 2.0 0.0 convergence_context = {}
1156
1157 # Perform the iterative optimization untill the final convergence
1158 # criterion
1159 37 164 4.4 0.0 for iteration_idx in xrange(n_iterations):
1160
1161 # Sample the minibatch from the full dataset
1162 37 185 5.0 0.0 minibatch_indices = self.random_state.random_integers(
1163 37 758 20.5 0.1 0, n_samples - 1, self.batch_size)
1164
1165 # Perform the actual update step on the minibatch data
1166 37 151 4.1 0.0 batch_inertia, centers_squared_diff = _mini_batch_step(
1167 37 120283 3250.9 23.4 X[minibatch_indices], x_squared_norms[minibatch_indices],
1168 37 216 5.8 0.0 self.cluster_centers_, self.counts_,
1169 37 299801 8102.7 58.4 old_center_buffer, tol > 0.0, distances=distances)
1170
1171 # Monitor the convergence and do early stopping if necessary
1172 37 193 5.2 0.0 if _mini_batch_convergence(
1173 37 148 4.0 0.0 self, iteration_idx, n_iterations, tol, n_samples,
1174 37 134 3.6 0.0 centers_squared_diff, batch_inertia, convergence_context,
1175 37 2374 64.2 0.5 verbose=self.verbose):
1176 1 4 4.0 0.0 break
1177
1178 1 5 5.0 0.0 if self.compute_labels:
1179 1 4 4.0 0.0 if self.verbose:
1180 print 'Computing label assignements and total inertia'
1181 1 4 4.0 0.0 self.labels_, self.inertia_ = _labels_inertia(
1182 1 1384 1384.0 0.3 X, x_squared_norms, self.cluster_centers_)
1183
1184 1 4 4.0 0.0 return self
MiniBatchKMeans-madelon¶
Benchmark setup
from sklearn.cluster import MiniBatchKMeans from deps import load_data kwargs = {'n_clusters': 9} X, y, X_t, y_t = load_data('madelon') obj = MiniBatchKMeans(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
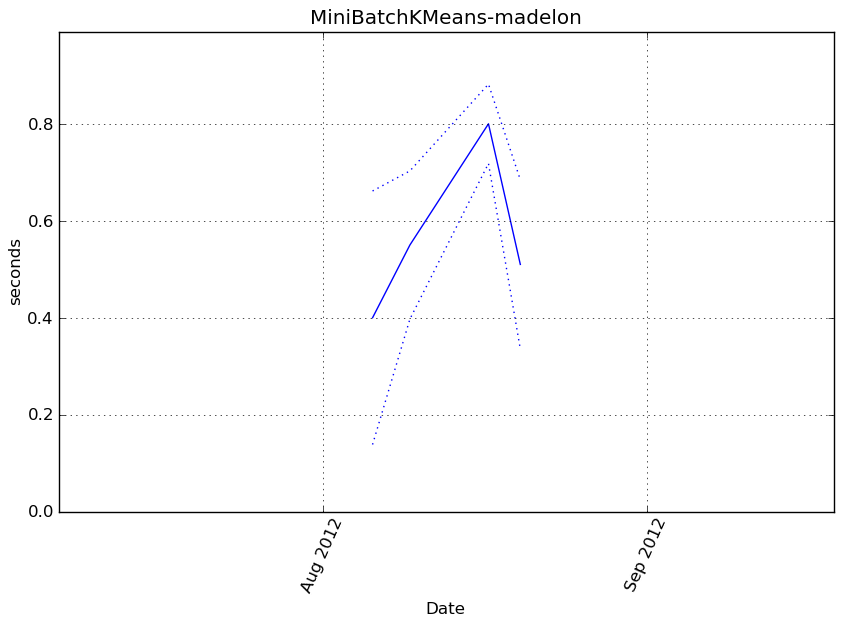
Memory usage
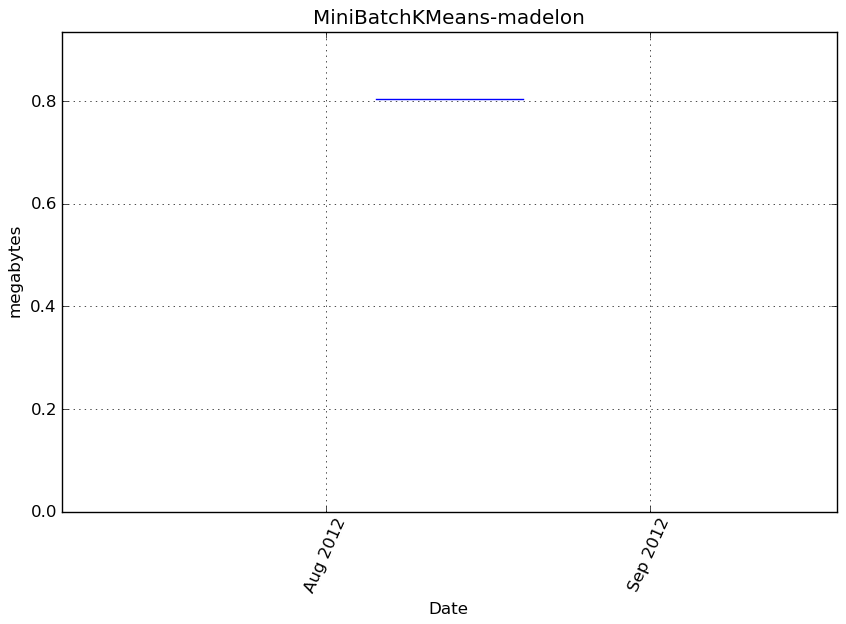
Additional output
cProfile
18745 function calls in 1.015 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 1.015 1.015 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 1.015 1.015 <f>:1(<module>)
1 0.258 0.258 1.015 1.015 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:1067(fit)
83 0.311 0.004 0.574 0.007 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:830(_mini_batch_step)
114 0.026 0.000 0.297 0.003 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:101(euclidean_distances)
87 0.002 0.000 0.260 0.003 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:431(_labels_inertia)
87 0.018 0.000 0.255 0.003 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:414(_labels_inertia_precompute_dense)
114 0.002 0.000 0.134 0.001 /tmp/vb_sklearn/sklearn/utils/extmath.py:70(safe_sparse_dot)
114 0.129 0.001 0.129 0.001 {numpy.core._dotblas.dot}
2365 0.112 0.000 0.112 0.000 {method 'sum' of 'numpy.ndarray' objects}
114 0.001 0.000 0.099 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
3 0.023 0.008 0.092 0.031 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:525(_init_centroids)
3 0.005 0.002 0.069 0.023 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:36(_k_init)
1456 0.002 0.000 0.067 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
1456 0.034 0.000 0.065 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
861 0.028 0.000 0.063 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
570 0.007 0.000 0.061 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
228 0.001 0.000 0.058 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
342 0.001 0.000 0.048 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
2329 0.029 0.000 0.029 0.000 {isinstance}
342 0.001 0.000 0.011 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
1 0.006 0.006 0.008 0.008 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:404(_squared_norms)
342 0.003 0.000 0.005 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
2912 0.003 0.000 0.003 0.000 {method 'split' of 'str' objects}
570 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
80 0.002 0.000 0.002 0.000 /tmp/vb_sklearn/sklearn/cluster/k_means_.py:898(_mini_batch_convergence)
913 0.002 0.000 0.002 0.000 {numpy.core.multiarray.array}
342 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
264 0.002 0.000 0.002 0.000 {numpy.core.multiarray.empty}
87 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
84 0.001 0.000 0.001 0.000 {method 'random_integers' of 'mtrand.RandomState' objects}
261 0.001 0.000 0.001 0.000 {method 'fill' of 'numpy.ndarray' objects}
1255 0.000 0.000 0.000 0.000 {len}
343 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
24 0.000 0.000 0.000 0.000 {method 'cumsum' of 'numpy.ndarray' objects}
24 0.000 0.000 0.000 0.000 {method 'random_sample' of 'mtrand.RandomState' objects}
171 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:94(check_arrays)
24 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:757(searchsorted)
320 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
24 0.000 0.000 0.000 0.000 {method 'searchsorted' of 'numpy.ndarray' objects}
7 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
9 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:289(ascontiguousarray)
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:87(_num_samples)
5 0.000 0.000 0.000 0.000 {hasattr}
3 0.000 0.000 0.000 0.000 {method 'randint' of 'mtrand.RandomState' objects}
4 0.000 0.000 0.000 0.000 {method 'pop' of 'dict' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/k_means_.py
Function: fit at line 1067
Total time: 0.788778 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
1067 def fit(self, X, y=None):
1068 """Compute the centroids on X by chunking it into mini-batches.
1069
1070 Parameters
1071 ----------
1072 X: array-like, shape = [n_samples, n_features]
1073 Coordinates of the data points to cluster
1074 """
1075 1 12 12.0 0.0 self.random_state = check_random_state(self.random_state)
1076 1 4 4.0 0.0 X = check_arrays(X, sparse_format="csr", copy=False,
1077 1 88 88.0 0.0 check_ccontiguous=True, dtype=np.float64)[0]
1078 1 5 5.0 0.0 n_samples, n_features = X.shape
1079 1 4 4.0 0.0 if n_samples < self.n_clusters:
1080 raise ValueError("Number of samples smaller than number "\
1081 "of clusters.")
1082
1083 1 7 7.0 0.0 if hasattr(self.init, '__array__'):
1084 self.init = np.ascontiguousarray(self.init, dtype=np.float64)
1085
1086 1 5350 5350.0 0.7 x_squared_norms = _squared_norms(X)
1087
1088 1 7 7.0 0.0 if self.tol > 0.0:
1089 tol = _tolerance(X, self.tol)
1090
1091 # using tol-based early stopping needs the allocation of a
1092 # dedicated before which can be expensive for high dim data:
1093 # hence we allocate it outside of the main loop
1094 old_center_buffer = np.zeros(n_features, np.double)
1095 else:
1096 1 3 3.0 0.0 tol = 0.0
1097 # no need for the center buffer if tol-based early stopping is
1098 # disabled
1099 1 12198 12198.0 1.5 old_center_buffer = np.zeros(0, np.double)
1100
1101 1 16 16.0 0.0 distances = np.zeros(self.batch_size, dtype=np.float64)
1102 1 37 37.0 0.0 n_batches = int(np.ceil(float(n_samples) / self.batch_size))
1103 1 5 5.0 0.0 n_iterations = int(self.max_iter * n_batches)
1104
1105 1 3 3.0 0.0 init_size = self.init_size
1106 1 4 4.0 0.0 if init_size is None:
1107 1 4 4.0 0.0 init_size = 3 * self.batch_size
1108 1 3 3.0 0.0 if init_size > n_samples:
1109 init_size = n_samples
1110 1 4 4.0 0.0 self.init_size_ = init_size
1111
1112 1 5 5.0 0.0 validation_indices = self.random_state.random_integers(
1113 1 24 24.0 0.0 0, n_samples - 1, init_size)
1114 1 7496 7496.0 1.0 X_valid = X[validation_indices]
1115 1 20 20.0 0.0 x_squared_norms_valid = x_squared_norms[validation_indices]
1116
1117 # perform several inits with random sub-sets
1118 1 4 4.0 0.0 best_inertia = None
1119 4 23 5.8 0.0 for init_idx in range(self.n_init):
1120 3 11 3.7 0.0 if self.verbose:
1121 print "Init %d/%d with method: %s" % (
1122 init_idx + 1, self.n_init, self.init)
1123 3 39 13.0 0.0 counts = np.zeros(self.n_clusters, dtype=np.int32)
1124
1125 # TODO: once the `k_means` function works with sparse input we
1126 # should refactor the following init to use it instead.
1127
1128 # Initialize the centers using only a fraction of the data as we
1129 # expect n_samples to be very large when using MiniBatchKMeans
1130 3 11 3.7 0.0 cluster_centers = _init_centroids(
1131 3 12 4.0 0.0 X, self.n_clusters, self.init,
1132 3 10 3.3 0.0 random_state=self.random_state,
1133 3 10 3.3 0.0 x_squared_norms=x_squared_norms,
1134 3 95051 31683.7 12.1 init_size=init_size)
1135
1136 # Compute the label assignement on the init dataset
1137 3 14 4.7 0.0 batch_inertia, centers_squared_diff = _mini_batch_step(
1138 3 55 18.3 0.0 X_valid, x_squared_norms[validation_indices],
1139 3 14 4.7 0.0 cluster_centers, counts, old_center_buffer, False,
1140 3 76115 25371.7 9.6 distances=distances)
1141
1142 # Keep only the best cluster centers across independent inits on
1143 # the common validation set
1144 3 15 5.0 0.0 _, inertia = _labels_inertia(X_valid, x_squared_norms_valid,
1145 3 22243 7414.3 2.8 cluster_centers)
1146 3 56 18.7 0.0 if self.verbose:
1147 print "Inertia for init %d/%d: %f" % (
1148 init_idx + 1, self.n_init, inertia)
1149 3 15 5.0 0.0 if best_inertia is None or inertia < best_inertia:
1150 2 12 6.0 0.0 self.cluster_centers_ = cluster_centers
1151 2 10 5.0 0.0 self.counts_ = counts
1152 2 7 3.5 0.0 best_inertia = inertia
1153
1154 # Empty context to be used inplace by the convergence check routine
1155 1 4 4.0 0.0 convergence_context = {}
1156
1157 # Perform the iterative optimization untill the final convergence
1158 # criterion
1159 46 212 4.6 0.0 for iteration_idx in xrange(n_iterations):
1160
1161 # Sample the minibatch from the full dataset
1162 46 217 4.7 0.0 minibatch_indices = self.random_state.random_integers(
1163 46 853 18.5 0.1 0, n_samples - 1, self.batch_size)
1164
1165 # Perform the actual update step on the minibatch data
1166 46 191 4.2 0.0 batch_inertia, centers_squared_diff = _mini_batch_step(
1167 46 134759 2929.5 17.1 X[minibatch_indices], x_squared_norms[minibatch_indices],
1168 46 585 12.7 0.1 self.cluster_centers_, self.counts_,
1169 46 369096 8023.8 46.8 old_center_buffer, tol > 0.0, distances=distances)
1170
1171 # Monitor the convergence and do early stopping if necessary
1172 46 304 6.6 0.0 if _mini_batch_convergence(
1173 46 181 3.9 0.0 self, iteration_idx, n_iterations, tol, n_samples,
1174 46 164 3.6 0.0 centers_squared_diff, batch_inertia, convergence_context,
1175 46 2794 60.7 0.4 verbose=self.verbose):
1176 1 5 5.0 0.0 break
1177
1178 1 4 4.0 0.0 if self.compute_labels:
1179 1 4 4.0 0.0 if self.verbose:
1180 print 'Computing label assignements and total inertia'
1181 1 4 4.0 0.0 self.labels_, self.inertia_ = _labels_inertia(
1182 1 60370 60370.0 7.7 X, x_squared_norms, self.cluster_centers_)
1183
1184 1 5 5.0 0.0 return self
Ward-minimadelon¶
Benchmark setup
from sklearn.cluster import Ward from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('minimadelon') obj = Ward(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
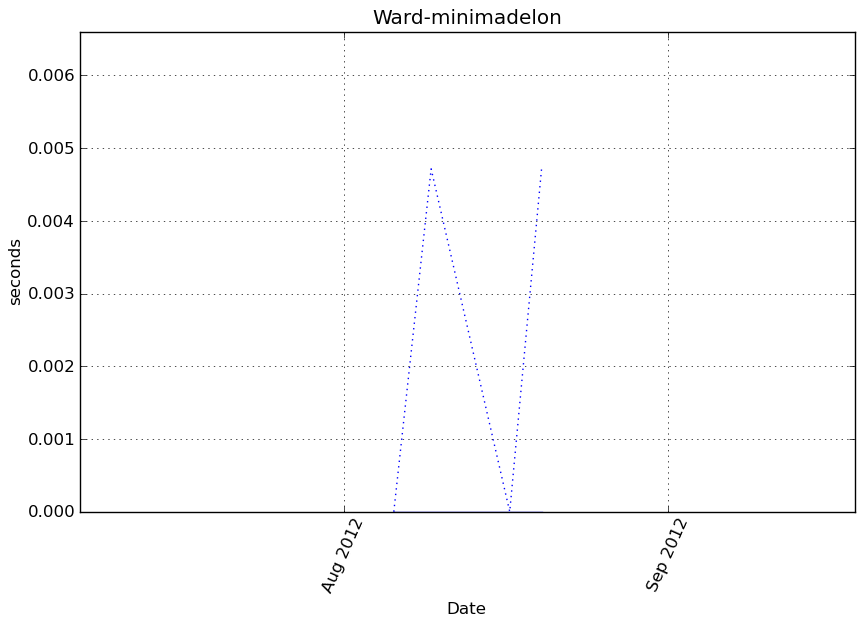
Memory usage
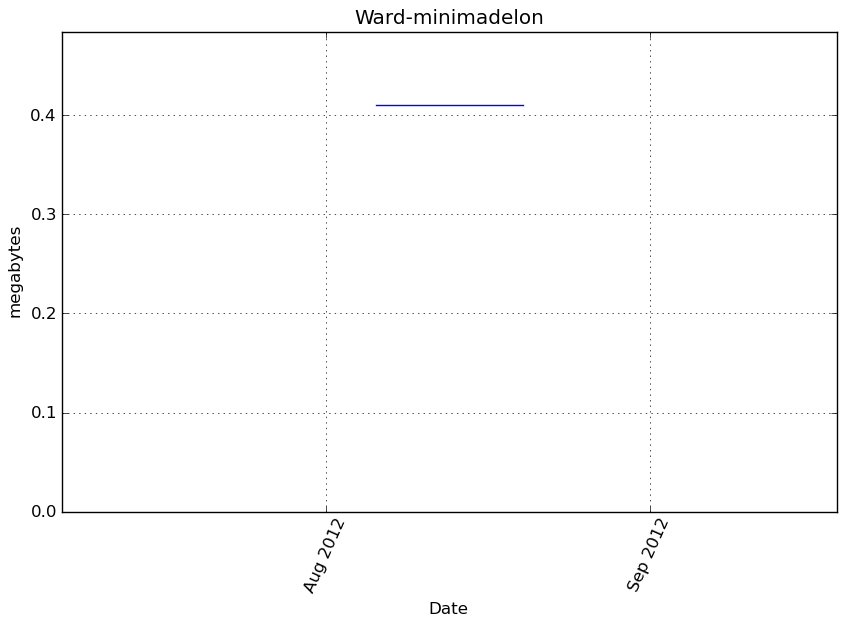
Additional output
cProfile
53 function calls in 0.002 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.002 0.002 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.002 0.002 <f>:1(<module>)
1 0.000 0.000 0.002 0.002 /tmp/vb_sklearn/sklearn/cluster/hierarchical.py:334(fit)
1 0.000 0.000 0.001 0.001 /tmp/vb_sklearn/sklearn/cluster/hierarchical.py:29(ward_tree)
1 0.000 0.000 0.001 0.001 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/cluster/hierarchy.py:423(ward)
1 0.000 0.000 0.001 0.001 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/cluster/hierarchy.py:464(linkage)
1 0.000 0.000 0.001 0.001 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:916(pdist)
1 0.001 0.001 0.001 0.001 {scipy.spatial._distance_wrap.pdist_euclidean_wrap}
1 0.000 0.000 0.000 0.000 {scipy.cluster._hierarchy_wrap.linkage_euclid_wrap}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/hierarchical.py:229(_hc_cut)
2 0.000 0.000 0.000 0.000 {sklearn.cluster._hierarchical._hc_get_descendent}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:132(_copy_arrays_if_base_present)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:120(_copy_array_if_base_present)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numerictypes.py:703(issubsctype)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numerictypes.py:608(obj2sctype)
3 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/cluster/hierarchy.py:912(_convert_to_double)
1 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {max}
3 0.000 0.000 0.000 0.000 {issubclass}
3 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
6 0.000 0.000 0.000 0.000 {isinstance}
1 0.000 0.000 0.000 0.000 {_heapq.heappush}
2 0.000 0.000 0.000 0.000 {method 'keys' of 'dict' objects}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:151(_convert_to_double)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/externals/joblib/memory.py:490(cache)
4 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 {_heapq.heappushpop}
1 0.000 0.000 0.000 0.000 {callable}
1 0.000 0.000 0.000 0.000 {method 'lower' of 'str' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/hierarchical.py
Function: fit at line 334
Total time: 0.00165 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
334 def fit(self, X):
335 """Fit the hierarchical clustering on the data
336
337 Parameters
338 ----------
339 X : array-like, shape = [n_samples, n_features]
340 The samples a.k.a. observations.
341
342 Returns
343 -------
344 self
345 """
346 1 4 4.0 0.2 memory = self.memory
347 1 6 6.0 0.4 if isinstance(memory, basestring):
348 memory = Memory(cachedir=memory)
349
350 1 3 3.0 0.2 if not self.connectivity is None:
351 if not sparse.issparse(self.connectivity):
352 raise TypeError("`connectivity` should be a sparse matrix or "
353 "None, got: %r" % type(self.connectivity))
354
355 if (self.connectivity.shape[0] != X.shape[0] or
356 self.connectivity.shape[1] != X.shape[0]):
357 raise ValueError("`connectivity` does not have shape "
358 "(n_samples, n_samples)")
359
360 1 4 4.0 0.2 n_samples = len(X)
361 1 3 3.0 0.2 compute_full_tree = self.compute_full_tree
362 1 3 3.0 0.2 if self.connectivity is None:
363 1 3 3.0 0.2 compute_full_tree = True
364 1 4 4.0 0.2 if compute_full_tree == 'auto':
365 # Early stopping is likely to give a speed up only for
366 # a large number of clusters. The actual threshold
367 # implemented here is heuristic
368 compute_full_tree = self.n_clusters > max(100, .02 * n_samples)
369 1 3 3.0 0.2 n_clusters = self.n_clusters
370 1 2 2.0 0.1 if compute_full_tree:
371 1 3 3.0 0.2 n_clusters = None
372
373 # Construct the tree
374 self.children_, self.n_components, self.n_leaves_, parents = \
375 1 10 10.0 0.6 memory.cache(ward_tree)(X, self.connectivity,
376 1 3 3.0 0.2 n_components=self.n_components,
377 1 1330 1330.0 80.6 copy=self.copy, n_clusters=n_clusters)
378 # Cut the tree
379 1 4 4.0 0.2 if compute_full_tree:
380 1 3 3.0 0.2 self.labels_ = _hc_cut(self.n_clusters, self.children_,
381 1 259 259.0 15.7 self.n_leaves_)
382 else:
383 labels = _hierarchical.hc_get_heads(parents, copy=False)
384 # copy to avoid holding a reference on the original array
385 labels = np.copy(labels[:n_samples])
386 # Reasign cluster numbers
387 self.labels_ = np.searchsorted(np.unique(labels), labels)
388 1 3 3.0 0.2 return self
Ward-blobs¶
Benchmark setup
from sklearn.cluster import Ward from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('blobs') obj = Ward(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
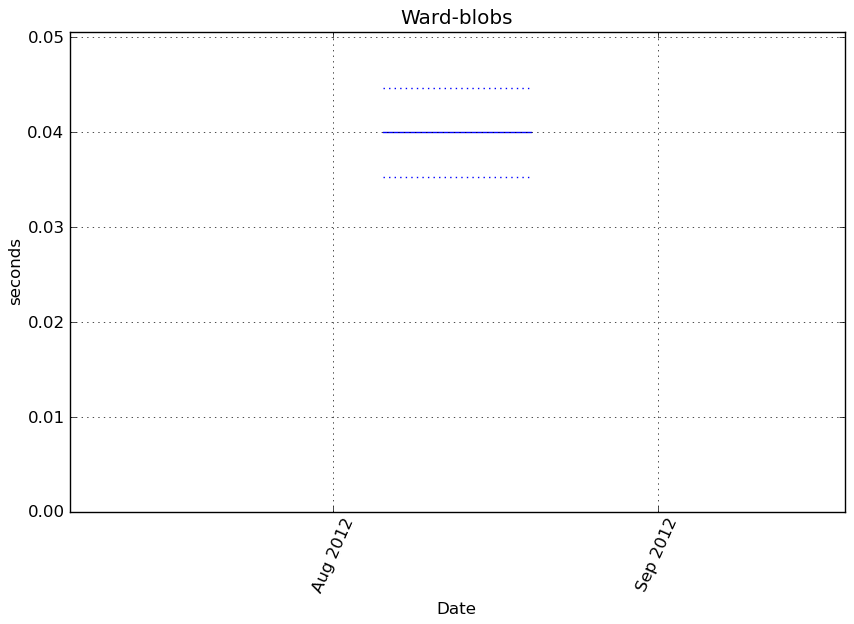
Memory usage
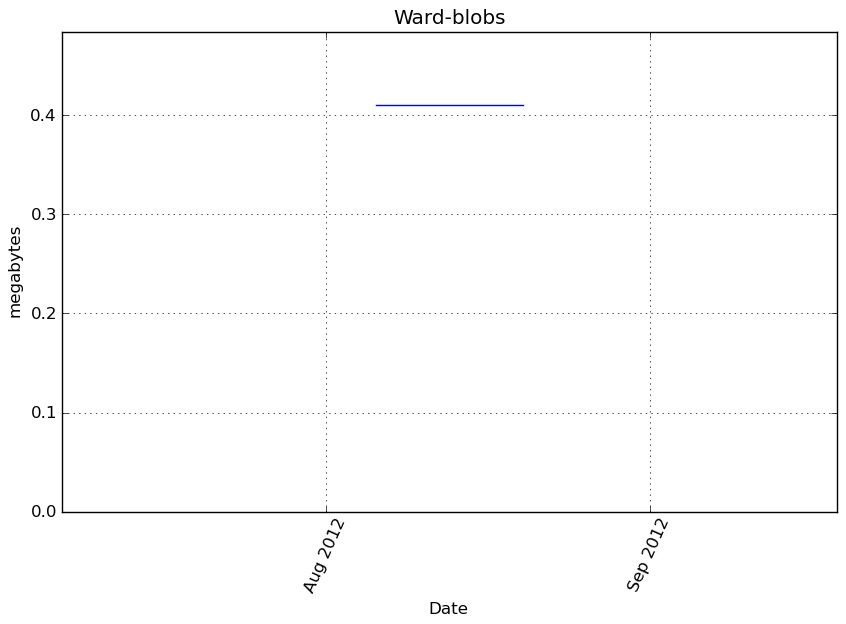
Additional output
cProfile
53 function calls in 0.046 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.046 0.046 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.046 0.046 <f>:1(<module>)
1 0.000 0.000 0.046 0.046 /tmp/vb_sklearn/sklearn/cluster/hierarchical.py:334(fit)
1 0.000 0.000 0.045 0.045 /tmp/vb_sklearn/sklearn/cluster/hierarchical.py:29(ward_tree)
1 0.000 0.000 0.045 0.045 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/cluster/hierarchy.py:423(ward)
1 0.000 0.000 0.045 0.045 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/cluster/hierarchy.py:464(linkage)
1 0.038 0.038 0.038 0.038 {scipy.cluster._hierarchy_wrap.linkage_euclid_wrap}
1 0.000 0.000 0.007 0.007 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:916(pdist)
1 0.006 0.006 0.006 0.006 {scipy.spatial._distance_wrap.pdist_euclidean_wrap}
1 0.000 0.000 0.001 0.001 /tmp/vb_sklearn/sklearn/cluster/hierarchical.py:229(_hc_cut)
2 0.001 0.000 0.001 0.000 {sklearn.cluster._hierarchical._hc_get_descendent}
3 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:132(_copy_arrays_if_base_present)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:120(_copy_array_if_base_present)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numerictypes.py:703(issubsctype)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numerictypes.py:608(obj2sctype)
1 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/cluster/hierarchy.py:912(_convert_to_double)
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 {max}
3 0.000 0.000 0.000 0.000 {issubclass}
3 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
6 0.000 0.000 0.000 0.000 {isinstance}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/spatial/distance.py:151(_convert_to_double)
1 0.000 0.000 0.000 0.000 {_heapq.heappush}
2 0.000 0.000 0.000 0.000 {method 'keys' of 'dict' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/externals/joblib/memory.py:490(cache)
4 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 {_heapq.heappushpop}
1 0.000 0.000 0.000 0.000 {callable}
1 0.000 0.000 0.000 0.000 {method 'lower' of 'str' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/hierarchical.py
Function: fit at line 334
Total time: 0.045278 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
334 def fit(self, X):
335 """Fit the hierarchical clustering on the data
336
337 Parameters
338 ----------
339 X : array-like, shape = [n_samples, n_features]
340 The samples a.k.a. observations.
341
342 Returns
343 -------
344 self
345 """
346 1 5 5.0 0.0 memory = self.memory
347 1 7 7.0 0.0 if isinstance(memory, basestring):
348 memory = Memory(cachedir=memory)
349
350 1 3 3.0 0.0 if not self.connectivity is None:
351 if not sparse.issparse(self.connectivity):
352 raise TypeError("`connectivity` should be a sparse matrix or "
353 "None, got: %r" % type(self.connectivity))
354
355 if (self.connectivity.shape[0] != X.shape[0] or
356 self.connectivity.shape[1] != X.shape[0]):
357 raise ValueError("`connectivity` does not have shape "
358 "(n_samples, n_samples)")
359
360 1 4 4.0 0.0 n_samples = len(X)
361 1 3 3.0 0.0 compute_full_tree = self.compute_full_tree
362 1 3 3.0 0.0 if self.connectivity is None:
363 1 3 3.0 0.0 compute_full_tree = True
364 1 4 4.0 0.0 if compute_full_tree == 'auto':
365 # Early stopping is likely to give a speed up only for
366 # a large number of clusters. The actual threshold
367 # implemented here is heuristic
368 compute_full_tree = self.n_clusters > max(100, .02 * n_samples)
369 1 3 3.0 0.0 n_clusters = self.n_clusters
370 1 3 3.0 0.0 if compute_full_tree:
371 1 2 2.0 0.0 n_clusters = None
372
373 # Construct the tree
374 self.children_, self.n_components, self.n_leaves_, parents = \
375 1 11 11.0 0.0 memory.cache(ward_tree)(X, self.connectivity,
376 1 3 3.0 0.0 n_components=self.n_components,
377 1 44146 44146.0 97.5 copy=self.copy, n_clusters=n_clusters)
378 # Cut the tree
379 1 5 5.0 0.0 if compute_full_tree:
380 1 4 4.0 0.0 self.labels_ = _hc_cut(self.n_clusters, self.children_,
381 1 1066 1066.0 2.4 self.n_leaves_)
382 else:
383 labels = _hierarchical.hc_get_heads(parents, copy=False)
384 # copy to avoid holding a reference on the original array
385 labels = np.copy(labels[:n_samples])
386 # Reasign cluster numbers
387 self.labels_ = np.searchsorted(np.unique(labels), labels)
388 1 3 3.0 0.0 return self
MeanShift-minimadelon¶
Benchmark setup
from sklearn.cluster import MeanShift from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('minimadelon') obj = MeanShift(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
Memory usage
Additional output
cProfile
7910 function calls in 0.139 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.139 0.139 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.139 0.139 <f>:1(<module>)
1 0.000 0.000 0.139 0.139 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:265(fit)
1 0.072 0.072 0.139 0.139 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:51(mean_shift)
114 0.001 0.000 0.046 0.000 /tmp/vb_sklearn/sklearn/neighbors/base.py:334(radius_neighbors)
124 0.001 0.000 0.024 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
236 0.001 0.000 0.022 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
111 0.013 0.000 0.020 0.000 {method 'query_radius' of 'sklearn.neighbors.ball_tree.BallTree' objects}
236 0.002 0.000 0.017 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
823 0.017 0.000 0.017 0.000 {numpy.core.multiarray.array}
347 0.001 0.000 0.015 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
112 0.000 0.000 0.010 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2299(mean)
112 0.010 0.000 0.010 0.000 {method 'mean' of 'numpy.ndarray' objects}
392 0.001 0.000 0.006 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
111 0.001 0.000 0.005 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:15(norm)
392 0.003 0.000 0.005 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
136 0.002 0.000 0.004 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
111 0.003 0.000 0.003 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/blas.py:30(get_blas_funcs)
2 0.000 0.000 0.003 0.001 /tmp/vb_sklearn/sklearn/neighbors/base.py:156(kneighbors)
1 0.000 0.000 0.002 0.002 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:16(estimate_bandwidth)
4 0.000 0.000 0.002 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:404(pairwise_distances)
4 0.000 0.000 0.002 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:101(euclidean_distances)
475 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
144 0.002 0.000 0.002 0.000 {method 'sum' of 'numpy.ndarray' objects}
111 0.001 0.000 0.002 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:6(atleast_1d)
1 0.001 0.001 0.001 0.001 {method 'query' of 'sklearn.neighbors.ball_tree.BallTree' objects}
784 0.001 0.000 0.001 0.000 {method 'split' of 'str' objects}
112 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
4 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
4 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/neighbors/base.py:578(fit)
4 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/neighbors/base.py:96(_fit)
12 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
524 0.001 0.000 0.001 0.000 {isinstance}
111 0.001 0.000 0.001 0.000 {method 'reshape' of 'numpy.ndarray' objects}
4 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:70(safe_sparse_dot)
112 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
1080 0.000 0.000 0.000 0.000 {len}
112 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
458 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
8 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
111 0.000 0.000 0.000 0.000 {getattr}
111 0.000 0.000 0.000 0.000 {max}
222 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/neighbors/unsupervised.py:85(__init__)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1774(amax)
1 0.000 0.000 0.000 0.000 {method 'max' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/neighbors/base.py:77(_init_params)
1 0.000 0.000 0.000 0.000 {sorted}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
1 0.000 0.000 0.000 0.000 {method 'any' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {method 'argsort' of 'numpy.ndarray' objects}
3 0.000 0.000 0.000 0.000 {numpy.core.multiarray.where}
2 0.000 0.000 0.000 0.000 {numpy.core.multiarray.arange}
1 0.000 0.000 0.000 0.000 {method 'flatten' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
6 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:134(<lambda>)
1 0.000 0.000 0.000 0.000 {method 'items' of 'dict' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py
Function: fit at line 265
Total time: 0.150272 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
265 def fit(self, X):
266 """ Compute MeanShift
267
268 Parameters
269 -----------
270 X : array [n_samples, n_features]
271 Input points
272 """
273 self.cluster_centers_, self.labels_ = \
274 1 3 3.0 0.0 mean_shift(X,
275 1 2 2.0 0.0 bandwidth=self.bandwidth,
276 1 2 2.0 0.0 seeds=self.seeds,
277 1 2 2.0 0.0 bin_seeding=self.bin_seeding,
278 1 150261 150261.0 100.0 cluster_all=self.cluster_all)
279 1 2 2.0 0.0 return self
MeanShift-blobs¶
Benchmark setup
from sklearn.cluster import MeanShift from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('blobs') obj = MeanShift(**kwargs)
Benchmark statement
obj.fit(X)
Execution time
Memory usage
Additional output
cProfile
73505 function calls in 1.452 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 1.452 1.452 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 1.452 1.452 <f>:1(<module>)
1 0.000 0.000 1.452 1.452 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:265(fit)
1 0.922 0.922 1.452 1.452 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:51(mean_shift)
1121 0.009 0.000 0.346 0.000 /tmp/vb_sklearn/sklearn/neighbors/base.py:334(radius_neighbors)
1120 0.114 0.000 0.189 0.000 {method 'query_radius' of 'sklearn.neighbors.ball_tree.BallTree' objects}
1127 0.006 0.000 0.148 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
2248 0.011 0.000 0.136 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
1121 0.003 0.000 0.087 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2299(mean)
1121 0.084 0.000 0.084 0.000 {method 'mean' of 'numpy.ndarray' objects}
3395 0.007 0.000 0.071 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
2248 0.015 0.000 0.065 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
3395 0.028 0.000 0.064 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
1120 0.021 0.000 0.060 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:15(norm)
7868 0.055 0.000 0.055 0.000 {numpy.core.multiarray.array}
3368 0.007 0.000 0.050 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
1 0.000 0.000 0.033 0.033 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:16(estimate_bandwidth)
1120 0.026 0.000 0.033 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/blas.py:30(get_blas_funcs)
2 0.000 0.000 0.032 0.016 /tmp/vb_sklearn/sklearn/neighbors/base.py:156(kneighbors)
6790 0.032 0.000 0.032 0.000 {method 'split' of 'str' objects}
1 0.031 0.031 0.031 0.031 {method 'query' of 'sklearn.neighbors.ball_tree.BallTree' objects}
1135 0.015 0.000 0.029 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
4499 0.008 0.000 0.020 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1120 0.007 0.000 0.018 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:6(atleast_1d)
1139 0.014 0.000 0.014 0.000 {method 'sum' of 'numpy.ndarray' objects}
1121 0.003 0.000 0.011 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
4532 0.007 0.000 0.007 0.000 {isinstance}
1120 0.006 0.000 0.006 0.000 {method 'reshape' of 'numpy.ndarray' objects}
1121 0.004 0.000 0.004 0.000 {method 'fill' of 'numpy.ndarray' objects}
10409 0.004 0.000 0.004 0.000 {len}
1121 0.003 0.000 0.003 0.000 {numpy.core.multiarray.empty}
4 0.000 0.000 0.003 0.001 /tmp/vb_sklearn/sklearn/neighbors/base.py:578(fit)
4 0.002 0.000 0.003 0.001 /tmp/vb_sklearn/sklearn/neighbors/base.py:96(_fit)
4488 0.002 0.000 0.002 0.000 {method 'append' of 'list' objects}
1120 0.002 0.000 0.002 0.000 {getattr}
2 0.000 0.000 0.001 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:404(pairwise_distances)
2 0.000 0.000 0.001 0.001 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:101(euclidean_distances)
1120 0.001 0.000 0.001 0.000 {max}
2240 0.001 0.000 0.001 0.000 {method 'get' of 'dict' objects}
8 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
2 0.000 0.000 0.001 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1774(amax)
1 0.000 0.000 0.000 0.000 {method 'max' of 'numpy.ndarray' objects}
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/extmath.py:70(safe_sparse_dot)
4 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/neighbors/unsupervised.py:85(__init__)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
2 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/neighbors/base.py:77(_init_params)
1 0.000 0.000 0.000 0.000 {method 'argsort' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {sorted}
1 0.000 0.000 0.000 0.000 {method 'flatten' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.arange}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.where}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py:134(<lambda>)
1 0.000 0.000 0.000 0.000 {method 'items' of 'dict' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/cluster/mean_shift_.py
Function: fit at line 265
Total time: 1.65172 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
265 def fit(self, X):
266 """ Compute MeanShift
267
268 Parameters
269 -----------
270 X : array [n_samples, n_features]
271 Input points
272 """
273 self.cluster_centers_, self.labels_ = \
274 1 4 4.0 0.0 mean_shift(X,
275 1 2 2.0 0.0 bandwidth=self.bandwidth,
276 1 1 1.0 0.0 seeds=self.seeds,
277 1 2 2.0 0.0 bin_seeding=self.bin_seeding,
278 1 1651711 1651711.0 100.0 cluster_all=self.cluster_all)
279 1 3 3.0 0.0 return self
