Benchmarks for gaussian_process¶
GaussianProcess-cobyla-blobs¶
Benchmark setup
from sklearn.gaussian_process import GaussianProcess from deps import load_data kwargs = {'optimizer': 'fmin_cobyla'} X, y, X_t, y_t = load_data('blobs') obj = GaussianProcess(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
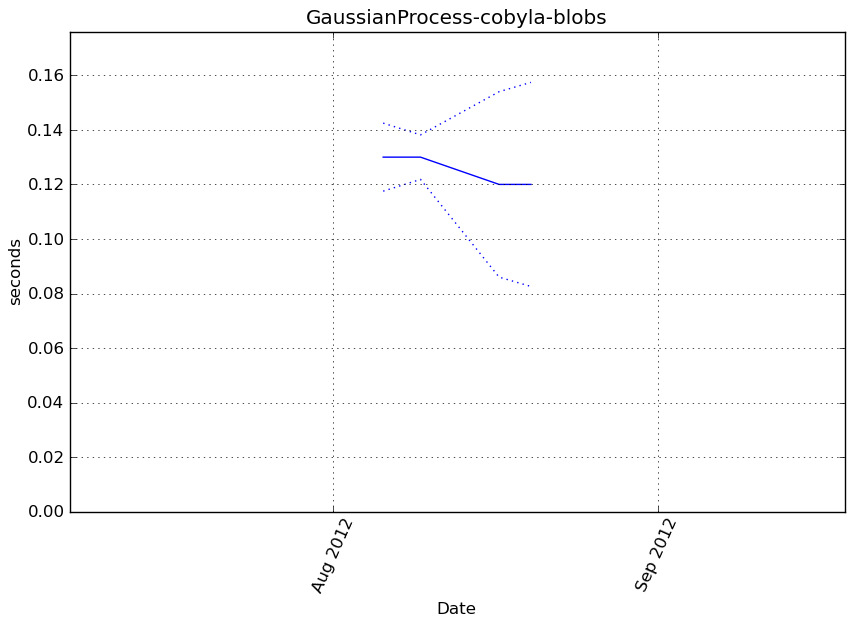
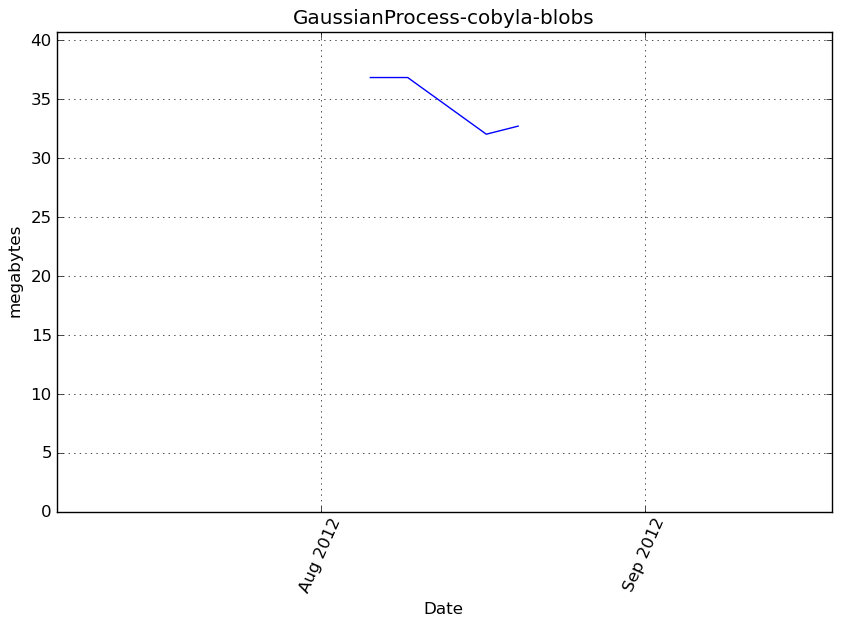
Memory usage
Additional output
cProfile
642 function calls in 0.143 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.143 0.143 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.143 0.143 <f>:1(<module>)
1 0.001 0.001 0.143 0.143 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:250(fit)
1 0.058 0.058 0.074 0.074 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:28(l1_cross_distances)
1 0.011 0.011 0.060 0.060 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:522(reduced_likelihood_function)
1 0.020 0.020 0.027 0.027 /tmp/vb_sklearn/sklearn/gaussian_process/correlation_models.py:58(squared_exponential)
2 0.000 0.000 0.015 0.007 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
4 0.015 0.004 0.015 0.004 {method 'sum' of 'numpy.ndarray' objects}
3 0.014 0.005 0.014 0.005 {numpy.core.multiarray.zeros}
11 0.010 0.001 0.012 0.001 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py:526(asarray_chkfinite)
4 0.002 0.000 0.011 0.003 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:72(solve_triangular)
1 0.000 0.000 0.010 0.010 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_cholesky.py:30(cholesky)
1 0.007 0.007 0.010 0.010 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_cholesky.py:13(_cholesky)
4 0.000 0.000 0.009 0.002 {map}
24 0.002 0.000 0.002 0.000 {method 'any' of 'numpy.ndarray' objects}
301 0.002 0.000 0.002 0.000 {numpy.core.multiarray.arange}
2 0.001 0.000 0.001 0.000 {method 'astype' of 'numpy.ndarray' objects}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2377(std)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_qr.py:16(qr)
8 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:60(get_lapack_funcs)
2 0.000 0.000 0.000 0.000 {method 'std' of 'numpy.ndarray' objects}
9 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:45(find_best_lapack_type)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_svd.py:14(svd)
3 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/twodim_base.py:169(eye)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1836(amin)
1 0.000 0.000 0.000 0.000 {method 'min' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:822(_check_params)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2299(mean)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/special_matrices.py:98(triu)
2 0.000 0.000 0.000 0.000 {method 'mean' of 'numpy.ndarray' objects}
21 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/special_matrices.py:20(tri)
24 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
22 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
13 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:23(cast_to_lapack_prefix)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/regression_models.py:16(constant)
10 0.000 0.000 0.000 0.000 {range}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/twodim_base.py:220(diag)
1 0.000 0.000 0.000 0.000 {method 'outer' of 'numpy.ufunc' objects}
12 0.000 0.000 0.000 0.000 {getattr}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
26 0.000 0.000 0.000 0.000 {issubclass}
2 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.generic' objects}
5 0.000 0.000 0.000 0.000 {isinstance}
9 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:449(isfortran)
2 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
24 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
27 0.000 0.000 0.000 0.000 {len}
9 0.000 0.000 0.000 0.000 {method 'sort' of 'list' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
1 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {method 'prod' of 'numpy.ndarray' objects}
7 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/misc.py:22(_datacopied)
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
1 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
2 0.000 0.000 0.000 0.000 {callable}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: fit at line 250
Total time: 0.143561 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
250 def fit(self, X, y):
251 """
252 The Gaussian Process model fitting method.
253
254 Parameters
255 ----------
256 X : double array_like
257 An array with shape (n_samples, n_features) with the input at which
258 observations were made.
259
260 y : double array_like
261 An array with shape (n_features, ) with the observations of the
262 scalar output to be predicted.
263
264 Returns
265 -------
266 gp : self
267 A fitted Gaussian Process model object awaiting data to perform
268 predictions.
269 """
270 1 13 13.0 0.0 self.random_state = check_random_state(self.random_state)
271
272 # Force data to 2D numpy.array
273 1 60 60.0 0.0 X = array2d(X)
274 1 20 20.0 0.0 y = np.asarray(y).ravel()[:, np.newaxis]
275
276 # Check shapes of DOE & observations
277 1 5 5.0 0.0 n_samples_X, n_features = X.shape
278 1 4 4.0 0.0 n_samples_y = y.shape[0]
279
280 1 4 4.0 0.0 if n_samples_X != n_samples_y:
281 raise ValueError("X and y must have the same number of rows.")
282 else:
283 1 4 4.0 0.0 n_samples = n_samples_X
284
285 # Run input checks
286 1 198 198.0 0.1 self._check_params(n_samples)
287
288 # Normalize data or don't
289 1 5 5.0 0.0 if self.normalize:
290 1 87 87.0 0.1 X_mean = np.mean(X, axis=0)
291 1 303 303.0 0.2 X_std = np.std(X, axis=0)
292 1 44 44.0 0.0 y_mean = np.mean(y, axis=0)
293 1 85 85.0 0.1 y_std = np.std(y, axis=0)
294 1 23 23.0 0.0 X_std[X_std == 0.] = 1.
295 1 17 17.0 0.0 y_std[y_std == 0.] = 1.
296 # center and scale X if necessary
297 1 396 396.0 0.3 X = (X - X_mean) / X_std
298 1 40 40.0 0.0 y = (y - y_mean) / y_std
299 else:
300 X_mean = np.zeros(1)
301 X_std = np.ones(1)
302 y_mean = np.zeros(1)
303 y_std = np.ones(1)
304
305 # Calculate matrix of distances D between samples
306 1 75002 75002.0 52.2 D, ij = l1_cross_distances(X)
307 1 7696 7696.0 5.4 if np.min(np.sum(D, axis=1)) == 0. \
308 and self.corr != correlation.pure_nugget:
309 raise Exception("Multiple input features cannot have the same"
310 " value")
311
312 # Regression matrix and parameters
313 1 52 52.0 0.0 F = self.regr(X)
314 1 5 5.0 0.0 n_samples_F = F.shape[0]
315 1 5 5.0 0.0 if F.ndim > 1:
316 1 5 5.0 0.0 p = F.shape[1]
317 else:
318 p = 1
319 1 4 4.0 0.0 if n_samples_F != n_samples:
320 raise Exception("Number of rows in F and X do not match. Most "
321 + "likely something is going wrong with the "
322 + "regression model.")
323 1 16 16.0 0.0 if p > n_samples_F:
324 raise Exception(("Ordinary least squares problem is undetermined "
325 + "n_samples=%d must be greater than the "
326 + "regression model size p=%d.") % (n_samples, p))
327 1 5 5.0 0.0 if self.beta0 is not None:
328 if self.beta0.shape[0] != p:
329 raise Exception("Shapes of beta0 and F do not match.")
330
331 # Set attributes
332 1 6 6.0 0.0 self.X = X
333 1 5 5.0 0.0 self.y = y
334 1 4 4.0 0.0 self.D = D
335 1 7 7.0 0.0 self.ij = ij
336 1 4 4.0 0.0 self.F = F
337 1 5 5.0 0.0 self.X_mean, self.X_std = X_mean, X_std
338 1 7 7.0 0.0 self.y_mean, self.y_std = y_mean, y_std
339
340 # Determine Gaussian Process model parameters
341 1 5 5.0 0.0 if self.thetaL is not None and self.thetaU is not None:
342 # Maximum Likelihood Estimation of the parameters
343 if self.verbose:
344 print("Performing Maximum Likelihood Estimation of the "
345 + "autocorrelation parameters...")
346 self.theta_, self.reduced_likelihood_function_value_, par = \
347 self._arg_max_reduced_likelihood_function()
348 if np.isinf(self.reduced_likelihood_function_value_):
349 raise Exception("Bad parameter region. "
350 + "Try increasing upper bound")
351
352 else:
353 # Given parameters
354 1 4 4.0 0.0 if self.verbose:
355 print("Given autocorrelation parameters. "
356 + "Computing Gaussian Process model parameters...")
357 1 6 6.0 0.0 self.theta_ = self.theta0
358 self.reduced_likelihood_function_value_, par = \
359 1 59343 59343.0 41.3 self.reduced_likelihood_function()
360 1 20 20.0 0.0 if np.isinf(self.reduced_likelihood_function_value_):
361 raise Exception("Bad point. Try increasing theta0.")
362
363 1 5 5.0 0.0 self.beta = par['beta']
364 1 5 5.0 0.0 self.gamma = par['gamma']
365 1 5 5.0 0.0 self.sigma2 = par['sigma2']
366 1 4 4.0 0.0 self.C = par['C']
367 1 4 4.0 0.0 self.Ft = par['Ft']
368 1 5 5.0 0.0 self.G = par['G']
369
370 1 15 15.0 0.0 if self.storage_mode == 'light':
371 # Delete heavy data (it will be computed again if required)
372 # (it is required only when MSE is wanted in self.predict)
373 if self.verbose:
374 print("Light storage mode specified. "
375 + "Flushing autocorrelation matrix...")
376 self.D = None
377 self.ij = None
378 self.F = None
379 self.C = None
380 self.Ft = None
381 self.G = None
382
383 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: predict at line 385
Total time: 0 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
385 def predict(self, X, eval_MSE=False, batch_size=None):
386 """
387 This function evaluates the Gaussian Process model at x.
388
389 Parameters
390 ----------
391 X : array_like
392 An array with shape (n_eval, n_features) giving the point(s) at
393 which the prediction(s) should be made.
394
395 eval_MSE : boolean, optional
396 A boolean specifying whether the Mean Squared Error should be
397 evaluated or not.
398 Default assumes evalMSE = False and evaluates only the BLUP (mean
399 prediction).
400
401 batch_size : integer, optional
402 An integer giving the maximum number of points that can be
403 evaluated simulatneously (depending on the available memory).
404 Default is None so that all given points are evaluated at the same
405 time.
406
407 Returns
408 -------
409 y : array_like
410 An array with shape (n_eval, ) with the Best Linear Unbiased
411 Prediction at x.
412
413 MSE : array_like, optional (if eval_MSE == True)
414 An array with shape (n_eval, ) with the Mean Squared Error at x.
415 """
416
417 # Check input shapes
418 X = array2d(X)
419 n_eval, n_features_X = X.shape
420 n_samples, n_features = self.X.shape
421
422 # Run input checks
423 self._check_params(n_samples)
424
425 if n_features_X != n_features:
426 raise ValueError(("The number of features in X (X.shape[1] = %d) "
427 + "should match the sample size used for fit() "
428 + "which is %d.") % (n_features_X, n_features))
429
430 if batch_size is None:
431 # No memory management
432 # (evaluates all given points in a single batch run)
433
434 # Normalize input
435 X = (X - self.X_mean) / self.X_std
436
437 # Initialize output
438 y = np.zeros(n_eval)
439 if eval_MSE:
440 MSE = np.zeros(n_eval)
441
442 # Get pairwise componentwise L1-distances to the input training set
443 dx = manhattan_distances(X, Y=self.X, sum_over_features=False)
444 # Get regression function and correlation
445 f = self.regr(X)
446 r = self.corr(self.theta_, dx).reshape(n_eval, n_samples)
447
448 # Scaled predictor
449 y_ = np.dot(f, self.beta) + np.dot(r, self.gamma)
450
451 # Predictor
452 y = (self.y_mean + self.y_std * y_).ravel()
453
454 # Mean Squared Error
455 if eval_MSE:
456 C = self.C
457 if C is None:
458 # Light storage mode (need to recompute C, F, Ft and G)
459 if self.verbose:
460 print("This GaussianProcess used 'light' storage mode "
461 + "at instanciation. Need to recompute "
462 + "autocorrelation matrix...")
463 reduced_likelihood_function_value, par = \
464 self.reduced_likelihood_function()
465 self.C = par['C']
466 self.Ft = par['Ft']
467 self.G = par['G']
468
469 rt = solve_triangular(self.C, r.T, lower=True)
470
471 if self.beta0 is None:
472 # Universal Kriging
473 u = solve_triangular(self.G.T,
474 np.dot(self.Ft.T, rt) - f.T)
475 else:
476 # Ordinary Kriging
477 u = np.zeros(y.shape)
478
479 MSE = self.sigma2 * (1. - (rt ** 2.).sum(axis=0)
480 + (u ** 2.).sum(axis=0))
481
482 # Mean Squared Error might be slightly negative depending on
483 # machine precision: force to zero!
484 MSE[MSE < 0.] = 0.
485
486 return y, MSE
487
488 else:
489
490 return y
491
492 else:
493 # Memory management
494
495 if type(batch_size) is not int or batch_size <= 0:
496 raise Exception("batch_size must be a positive integer")
497
498 if eval_MSE:
499
500 y, MSE = np.zeros(n_eval), np.zeros(n_eval)
501 for k in range(max(1, n_eval / batch_size)):
502 batch_from = k * batch_size
503 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
504 y[batch_from:batch_to], MSE[batch_from:batch_to] = \
505 self.predict(X[batch_from:batch_to],
506 eval_MSE=eval_MSE, batch_size=None)
507
508 return y, MSE
509
510 else:
511
512 y = np.zeros(n_eval)
513 for k in range(max(1, n_eval / batch_size)):
514 batch_from = k * batch_size
515 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
516 y[batch_from:batch_to] = \
517 self.predict(X[batch_from:batch_to],
518 eval_MSE=eval_MSE, batch_size=None)
519
520 return y
Benchmark statement
obj.predict(X_t)
Execution time
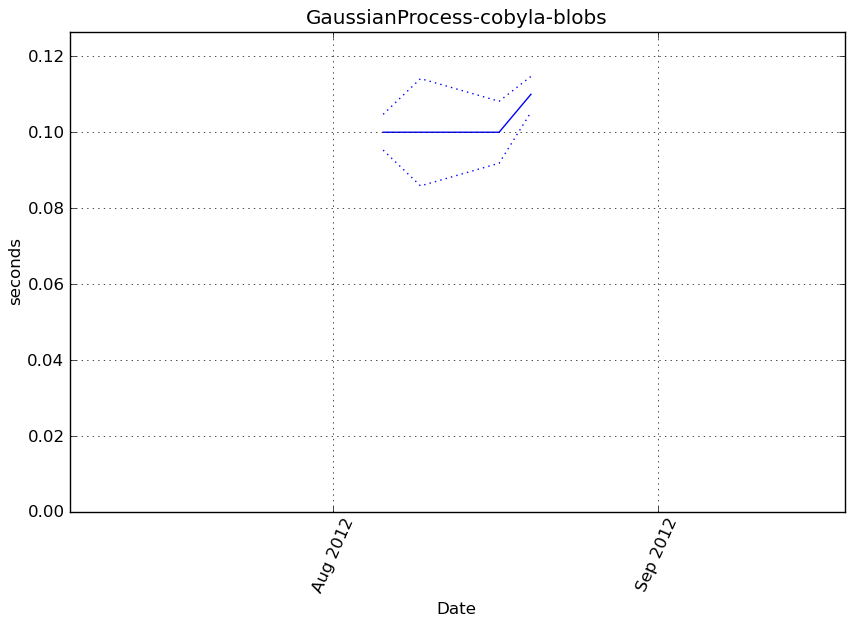
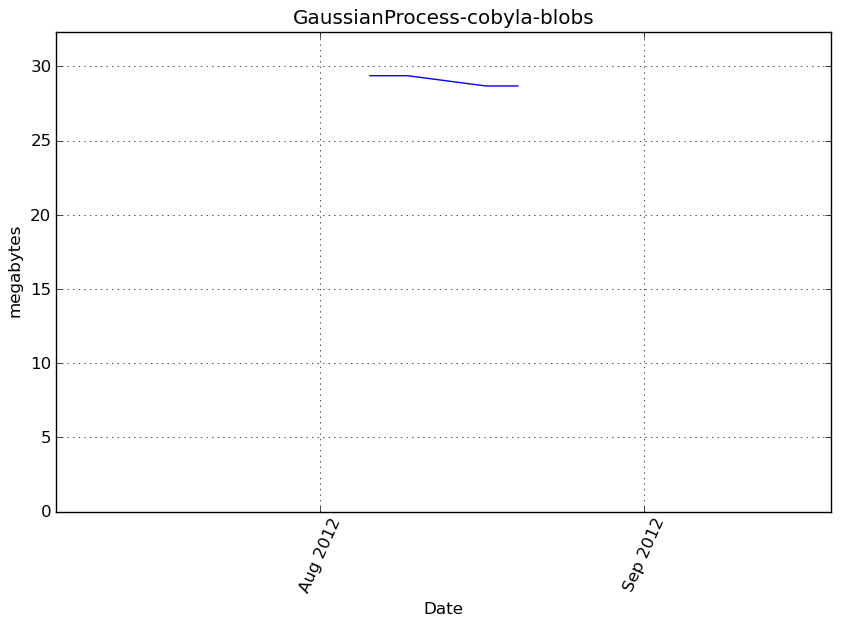
Memory usage
Additional output
cProfile
133 function calls in 0.100 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.100 0.100 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.100 0.100 <f>:1(<module>)
1 0.000 0.000 0.100 0.100 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:385(predict)
1 0.075 0.075 0.075 0.075 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:188(manhattan_distances)
1 0.014 0.014 0.024 0.024 /tmp/vb_sklearn/sklearn/gaussian_process/correlation_models.py:58(squared_exponential)
5 0.010 0.002 0.010 0.002 {method 'sum' of 'numpy.ndarray' objects}
1 0.000 0.000 0.010 0.010 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:822(_check_params)
8 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
2 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
8 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
4 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/regression_models.py:16(constant)
10 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
14 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
2 0.000 0.000 0.000 0.000 {method 'any' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
2 0.000 0.000 0.000 0.000 {method 'reshape' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
9 0.000 0.000 0.000 0.000 {isinstance}
16 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
14 0.000 0.000 0.000 0.000 {len}
4 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
1 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
2 0.000 0.000 0.000 0.000 {callable}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: fit at line 250
Total time: 0.143561 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
250 def fit(self, X, y):
251 """
252 The Gaussian Process model fitting method.
253
254 Parameters
255 ----------
256 X : double array_like
257 An array with shape (n_samples, n_features) with the input at which
258 observations were made.
259
260 y : double array_like
261 An array with shape (n_features, ) with the observations of the
262 scalar output to be predicted.
263
264 Returns
265 -------
266 gp : self
267 A fitted Gaussian Process model object awaiting data to perform
268 predictions.
269 """
270 1 13 13.0 0.0 self.random_state = check_random_state(self.random_state)
271
272 # Force data to 2D numpy.array
273 1 60 60.0 0.0 X = array2d(X)
274 1 20 20.0 0.0 y = np.asarray(y).ravel()[:, np.newaxis]
275
276 # Check shapes of DOE & observations
277 1 5 5.0 0.0 n_samples_X, n_features = X.shape
278 1 4 4.0 0.0 n_samples_y = y.shape[0]
279
280 1 4 4.0 0.0 if n_samples_X != n_samples_y:
281 raise ValueError("X and y must have the same number of rows.")
282 else:
283 1 4 4.0 0.0 n_samples = n_samples_X
284
285 # Run input checks
286 1 198 198.0 0.1 self._check_params(n_samples)
287
288 # Normalize data or don't
289 1 5 5.0 0.0 if self.normalize:
290 1 87 87.0 0.1 X_mean = np.mean(X, axis=0)
291 1 303 303.0 0.2 X_std = np.std(X, axis=0)
292 1 44 44.0 0.0 y_mean = np.mean(y, axis=0)
293 1 85 85.0 0.1 y_std = np.std(y, axis=0)
294 1 23 23.0 0.0 X_std[X_std == 0.] = 1.
295 1 17 17.0 0.0 y_std[y_std == 0.] = 1.
296 # center and scale X if necessary
297 1 396 396.0 0.3 X = (X - X_mean) / X_std
298 1 40 40.0 0.0 y = (y - y_mean) / y_std
299 else:
300 X_mean = np.zeros(1)
301 X_std = np.ones(1)
302 y_mean = np.zeros(1)
303 y_std = np.ones(1)
304
305 # Calculate matrix of distances D between samples
306 1 75002 75002.0 52.2 D, ij = l1_cross_distances(X)
307 1 7696 7696.0 5.4 if np.min(np.sum(D, axis=1)) == 0. \
308 and self.corr != correlation.pure_nugget:
309 raise Exception("Multiple input features cannot have the same"
310 " value")
311
312 # Regression matrix and parameters
313 1 52 52.0 0.0 F = self.regr(X)
314 1 5 5.0 0.0 n_samples_F = F.shape[0]
315 1 5 5.0 0.0 if F.ndim > 1:
316 1 5 5.0 0.0 p = F.shape[1]
317 else:
318 p = 1
319 1 4 4.0 0.0 if n_samples_F != n_samples:
320 raise Exception("Number of rows in F and X do not match. Most "
321 + "likely something is going wrong with the "
322 + "regression model.")
323 1 16 16.0 0.0 if p > n_samples_F:
324 raise Exception(("Ordinary least squares problem is undetermined "
325 + "n_samples=%d must be greater than the "
326 + "regression model size p=%d.") % (n_samples, p))
327 1 5 5.0 0.0 if self.beta0 is not None:
328 if self.beta0.shape[0] != p:
329 raise Exception("Shapes of beta0 and F do not match.")
330
331 # Set attributes
332 1 6 6.0 0.0 self.X = X
333 1 5 5.0 0.0 self.y = y
334 1 4 4.0 0.0 self.D = D
335 1 7 7.0 0.0 self.ij = ij
336 1 4 4.0 0.0 self.F = F
337 1 5 5.0 0.0 self.X_mean, self.X_std = X_mean, X_std
338 1 7 7.0 0.0 self.y_mean, self.y_std = y_mean, y_std
339
340 # Determine Gaussian Process model parameters
341 1 5 5.0 0.0 if self.thetaL is not None and self.thetaU is not None:
342 # Maximum Likelihood Estimation of the parameters
343 if self.verbose:
344 print("Performing Maximum Likelihood Estimation of the "
345 + "autocorrelation parameters...")
346 self.theta_, self.reduced_likelihood_function_value_, par = \
347 self._arg_max_reduced_likelihood_function()
348 if np.isinf(self.reduced_likelihood_function_value_):
349 raise Exception("Bad parameter region. "
350 + "Try increasing upper bound")
351
352 else:
353 # Given parameters
354 1 4 4.0 0.0 if self.verbose:
355 print("Given autocorrelation parameters. "
356 + "Computing Gaussian Process model parameters...")
357 1 6 6.0 0.0 self.theta_ = self.theta0
358 self.reduced_likelihood_function_value_, par = \
359 1 59343 59343.0 41.3 self.reduced_likelihood_function()
360 1 20 20.0 0.0 if np.isinf(self.reduced_likelihood_function_value_):
361 raise Exception("Bad point. Try increasing theta0.")
362
363 1 5 5.0 0.0 self.beta = par['beta']
364 1 5 5.0 0.0 self.gamma = par['gamma']
365 1 5 5.0 0.0 self.sigma2 = par['sigma2']
366 1 4 4.0 0.0 self.C = par['C']
367 1 4 4.0 0.0 self.Ft = par['Ft']
368 1 5 5.0 0.0 self.G = par['G']
369
370 1 15 15.0 0.0 if self.storage_mode == 'light':
371 # Delete heavy data (it will be computed again if required)
372 # (it is required only when MSE is wanted in self.predict)
373 if self.verbose:
374 print("Light storage mode specified. "
375 + "Flushing autocorrelation matrix...")
376 self.D = None
377 self.ij = None
378 self.F = None
379 self.C = None
380 self.Ft = None
381 self.G = None
382
383 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: predict at line 385
Total time: 0.099075 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
385 def predict(self, X, eval_MSE=False, batch_size=None):
386 """
387 This function evaluates the Gaussian Process model at x.
388
389 Parameters
390 ----------
391 X : array_like
392 An array with shape (n_eval, n_features) giving the point(s) at
393 which the prediction(s) should be made.
394
395 eval_MSE : boolean, optional
396 A boolean specifying whether the Mean Squared Error should be
397 evaluated or not.
398 Default assumes evalMSE = False and evaluates only the BLUP (mean
399 prediction).
400
401 batch_size : integer, optional
402 An integer giving the maximum number of points that can be
403 evaluated simulatneously (depending on the available memory).
404 Default is None so that all given points are evaluated at the same
405 time.
406
407 Returns
408 -------
409 y : array_like
410 An array with shape (n_eval, ) with the Best Linear Unbiased
411 Prediction at x.
412
413 MSE : array_like, optional (if eval_MSE == True)
414 An array with shape (n_eval, ) with the Mean Squared Error at x.
415 """
416
417 # Check input shapes
418 1 69 69.0 0.1 X = array2d(X)
419 1 6 6.0 0.0 n_eval, n_features_X = X.shape
420 1 5 5.0 0.0 n_samples, n_features = self.X.shape
421
422 # Run input checks
423 1 187 187.0 0.2 self._check_params(n_samples)
424
425 1 4 4.0 0.0 if n_features_X != n_features:
426 raise ValueError(("The number of features in X (X.shape[1] = %d) "
427 + "should match the sample size used for fit() "
428 + "which is %d.") % (n_features_X, n_features))
429
430 1 4 4.0 0.0 if batch_size is None:
431 # No memory management
432 # (evaluates all given points in a single batch run)
433
434 # Normalize input
435 1 287 287.0 0.3 X = (X - self.X_mean) / self.X_std
436
437 # Initialize output
438 1 15 15.0 0.0 y = np.zeros(n_eval)
439 1 4 4.0 0.0 if eval_MSE:
440 MSE = np.zeros(n_eval)
441
442 # Get pairwise componentwise L1-distances to the input training set
443 1 74459 74459.0 75.2 dx = manhattan_distances(X, Y=self.X, sum_over_features=False)
444 # Get regression function and correlation
445 1 57 57.0 0.1 f = self.regr(X)
446 1 23786 23786.0 24.0 r = self.corr(self.theta_, dx).reshape(n_eval, n_samples)
447
448 # Scaled predictor
449 1 144 144.0 0.1 y_ = np.dot(f, self.beta) + np.dot(r, self.gamma)
450
451 # Predictor
452 1 39 39.0 0.0 y = (self.y_mean + self.y_std * y_).ravel()
453
454 # Mean Squared Error
455 1 5 5.0 0.0 if eval_MSE:
456 C = self.C
457 if C is None:
458 # Light storage mode (need to recompute C, F, Ft and G)
459 if self.verbose:
460 print("This GaussianProcess used 'light' storage mode "
461 + "at instanciation. Need to recompute "
462 + "autocorrelation matrix...")
463 reduced_likelihood_function_value, par = \
464 self.reduced_likelihood_function()
465 self.C = par['C']
466 self.Ft = par['Ft']
467 self.G = par['G']
468
469 rt = solve_triangular(self.C, r.T, lower=True)
470
471 if self.beta0 is None:
472 # Universal Kriging
473 u = solve_triangular(self.G.T,
474 np.dot(self.Ft.T, rt) - f.T)
475 else:
476 # Ordinary Kriging
477 u = np.zeros(y.shape)
478
479 MSE = self.sigma2 * (1. - (rt ** 2.).sum(axis=0)
480 + (u ** 2.).sum(axis=0))
481
482 # Mean Squared Error might be slightly negative depending on
483 # machine precision: force to zero!
484 MSE[MSE < 0.] = 0.
485
486 return y, MSE
487
488 else:
489
490 1 4 4.0 0.0 return y
491
492 else:
493 # Memory management
494
495 if type(batch_size) is not int or batch_size <= 0:
496 raise Exception("batch_size must be a positive integer")
497
498 if eval_MSE:
499
500 y, MSE = np.zeros(n_eval), np.zeros(n_eval)
501 for k in range(max(1, n_eval / batch_size)):
502 batch_from = k * batch_size
503 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
504 y[batch_from:batch_to], MSE[batch_from:batch_to] = \
505 self.predict(X[batch_from:batch_to],
506 eval_MSE=eval_MSE, batch_size=None)
507
508 return y, MSE
509
510 else:
511
512 y = np.zeros(n_eval)
513 for k in range(max(1, n_eval / batch_size)):
514 batch_from = k * batch_size
515 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
516 y[batch_from:batch_to] = \
517 self.predict(X[batch_from:batch_to],
518 eval_MSE=eval_MSE, batch_size=None)
519
520 return y
GaussianProcess-Welch-blobs¶
Benchmark setup
from sklearn.gaussian_process import GaussianProcess from deps import load_data kwargs = {'optimizer': 'Welch'} X, y, X_t, y_t = load_data('blobs') obj = GaussianProcess(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
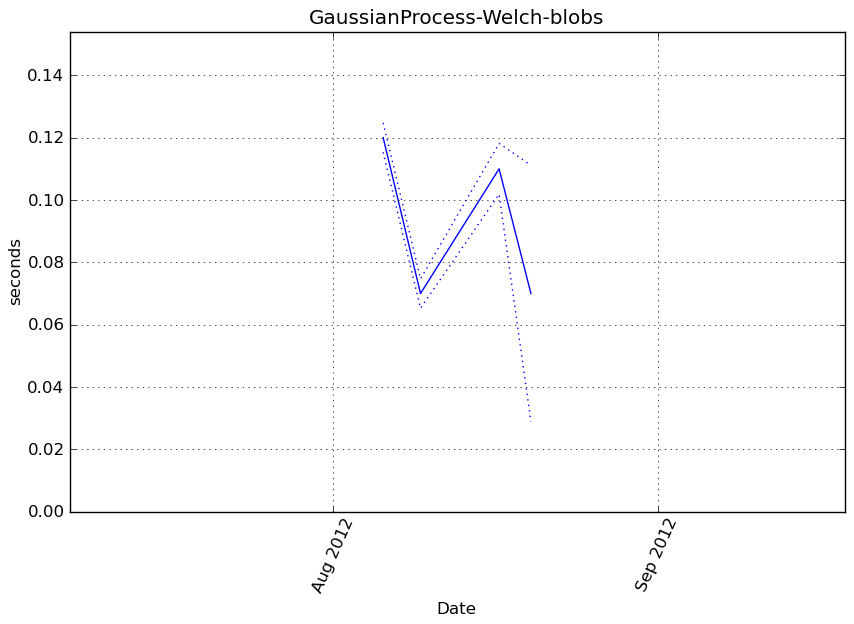
Memory usage
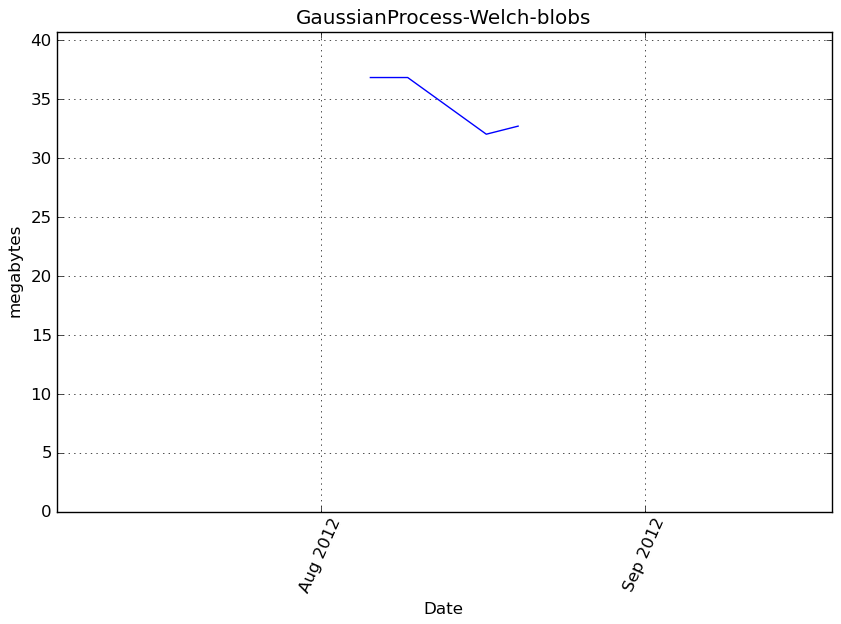
Additional output
cProfile
642 function calls in 0.142 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.142 0.142 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.142 0.142 <f>:1(<module>)
1 0.001 0.001 0.142 0.142 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:250(fit)
1 0.058 0.058 0.073 0.073 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:28(l1_cross_distances)
1 0.011 0.011 0.060 0.060 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:522(reduced_likelihood_function)
1 0.019 0.019 0.026 0.026 /tmp/vb_sklearn/sklearn/gaussian_process/correlation_models.py:58(squared_exponential)
2 0.000 0.000 0.015 0.007 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
4 0.015 0.004 0.015 0.004 {method 'sum' of 'numpy.ndarray' objects}
3 0.013 0.004 0.013 0.004 {numpy.core.multiarray.zeros}
11 0.010 0.001 0.012 0.001 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py:526(asarray_chkfinite)
4 0.002 0.000 0.011 0.003 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:72(solve_triangular)
1 0.000 0.000 0.010 0.010 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_cholesky.py:30(cholesky)
1 0.007 0.007 0.010 0.010 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_cholesky.py:13(_cholesky)
4 0.000 0.000 0.009 0.002 {map}
24 0.002 0.000 0.002 0.000 {method 'any' of 'numpy.ndarray' objects}
301 0.002 0.000 0.002 0.000 {numpy.core.multiarray.arange}
8 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:60(get_lapack_funcs)
2 0.001 0.000 0.001 0.000 {method 'astype' of 'numpy.ndarray' objects}
2 0.000 0.000 0.001 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2377(std)
2 0.001 0.000 0.001 0.000 {method 'std' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_qr.py:16(qr)
9 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:45(find_best_lapack_type)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1836(amin)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/decomp_svd.py:14(svd)
1 0.000 0.000 0.000 0.000 {method 'min' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/twodim_base.py:169(eye)
3 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:822(_check_params)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:2299(mean)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/special_matrices.py:98(triu)
2 0.000 0.000 0.000 0.000 {method 'mean' of 'numpy.ndarray' objects}
21 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/special_matrices.py:20(tri)
24 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
13 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:23(cast_to_lapack_prefix)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/regression_models.py:16(constant)
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
22 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
10 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/twodim_base.py:220(diag)
12 0.000 0.000 0.000 0.000 {getattr}
1 0.000 0.000 0.000 0.000 {method 'outer' of 'numpy.ufunc' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
26 0.000 0.000 0.000 0.000 {issubclass}
9 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:449(isfortran)
5 0.000 0.000 0.000 0.000 {isinstance}
3 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
2 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.generic' objects}
2 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
24 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
9 0.000 0.000 0.000 0.000 {method 'sort' of 'list' objects}
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:181(check_random_state)
27 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {method 'prod' of 'numpy.ndarray' objects}
7 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/misc.py:22(_datacopied)
1 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
4 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
2 0.000 0.000 0.000 0.000 {callable}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: fit at line 250
Total time: 0.143732 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
250 def fit(self, X, y):
251 """
252 The Gaussian Process model fitting method.
253
254 Parameters
255 ----------
256 X : double array_like
257 An array with shape (n_samples, n_features) with the input at which
258 observations were made.
259
260 y : double array_like
261 An array with shape (n_features, ) with the observations of the
262 scalar output to be predicted.
263
264 Returns
265 -------
266 gp : self
267 A fitted Gaussian Process model object awaiting data to perform
268 predictions.
269 """
270 1 13 13.0 0.0 self.random_state = check_random_state(self.random_state)
271
272 # Force data to 2D numpy.array
273 1 61 61.0 0.0 X = array2d(X)
274 1 20 20.0 0.0 y = np.asarray(y).ravel()[:, np.newaxis]
275
276 # Check shapes of DOE & observations
277 1 5 5.0 0.0 n_samples_X, n_features = X.shape
278 1 5 5.0 0.0 n_samples_y = y.shape[0]
279
280 1 4 4.0 0.0 if n_samples_X != n_samples_y:
281 raise ValueError("X and y must have the same number of rows.")
282 else:
283 1 4 4.0 0.0 n_samples = n_samples_X
284
285 # Run input checks
286 1 188 188.0 0.1 self._check_params(n_samples)
287
288 # Normalize data or don't
289 1 5 5.0 0.0 if self.normalize:
290 1 89 89.0 0.1 X_mean = np.mean(X, axis=0)
291 1 280 280.0 0.2 X_std = np.std(X, axis=0)
292 1 44 44.0 0.0 y_mean = np.mean(y, axis=0)
293 1 85 85.0 0.1 y_std = np.std(y, axis=0)
294 1 23 23.0 0.0 X_std[X_std == 0.] = 1.
295 1 17 17.0 0.0 y_std[y_std == 0.] = 1.
296 # center and scale X if necessary
297 1 404 404.0 0.3 X = (X - X_mean) / X_std
298 1 42 42.0 0.0 y = (y - y_mean) / y_std
299 else:
300 X_mean = np.zeros(1)
301 X_std = np.ones(1)
302 y_mean = np.zeros(1)
303 y_std = np.ones(1)
304
305 # Calculate matrix of distances D between samples
306 1 74826 74826.0 52.1 D, ij = l1_cross_distances(X)
307 1 7645 7645.0 5.3 if np.min(np.sum(D, axis=1)) == 0. \
308 and self.corr != correlation.pure_nugget:
309 raise Exception("Multiple input features cannot have the same"
310 " value")
311
312 # Regression matrix and parameters
313 1 51 51.0 0.0 F = self.regr(X)
314 1 5 5.0 0.0 n_samples_F = F.shape[0]
315 1 6 6.0 0.0 if F.ndim > 1:
316 1 5 5.0 0.0 p = F.shape[1]
317 else:
318 p = 1
319 1 5 5.0 0.0 if n_samples_F != n_samples:
320 raise Exception("Number of rows in F and X do not match. Most "
321 + "likely something is going wrong with the "
322 + "regression model.")
323 1 4 4.0 0.0 if p > n_samples_F:
324 raise Exception(("Ordinary least squares problem is undetermined "
325 + "n_samples=%d must be greater than the "
326 + "regression model size p=%d.") % (n_samples, p))
327 1 5 5.0 0.0 if self.beta0 is not None:
328 if self.beta0.shape[0] != p:
329 raise Exception("Shapes of beta0 and F do not match.")
330
331 # Set attributes
332 1 6 6.0 0.0 self.X = X
333 1 4 4.0 0.0 self.y = y
334 1 6 6.0 0.0 self.D = D
335 1 4 4.0 0.0 self.ij = ij
336 1 5 5.0 0.0 self.F = F
337 1 5 5.0 0.0 self.X_mean, self.X_std = X_mean, X_std
338 1 7 7.0 0.0 self.y_mean, self.y_std = y_mean, y_std
339
340 # Determine Gaussian Process model parameters
341 1 6 6.0 0.0 if self.thetaL is not None and self.thetaU is not None:
342 # Maximum Likelihood Estimation of the parameters
343 if self.verbose:
344 print("Performing Maximum Likelihood Estimation of the "
345 + "autocorrelation parameters...")
346 self.theta_, self.reduced_likelihood_function_value_, par = \
347 self._arg_max_reduced_likelihood_function()
348 if np.isinf(self.reduced_likelihood_function_value_):
349 raise Exception("Bad parameter region. "
350 + "Try increasing upper bound")
351
352 else:
353 # Given parameters
354 1 4 4.0 0.0 if self.verbose:
355 print("Given autocorrelation parameters. "
356 + "Computing Gaussian Process model parameters...")
357 1 5 5.0 0.0 self.theta_ = self.theta0
358 self.reduced_likelihood_function_value_, par = \
359 1 59780 59780.0 41.6 self.reduced_likelihood_function()
360 1 21 21.0 0.0 if np.isinf(self.reduced_likelihood_function_value_):
361 raise Exception("Bad point. Try increasing theta0.")
362
363 1 5 5.0 0.0 self.beta = par['beta']
364 1 5 5.0 0.0 self.gamma = par['gamma']
365 1 5 5.0 0.0 self.sigma2 = par['sigma2']
366 1 5 5.0 0.0 self.C = par['C']
367 1 5 5.0 0.0 self.Ft = par['Ft']
368 1 4 4.0 0.0 self.G = par['G']
369
370 1 5 5.0 0.0 if self.storage_mode == 'light':
371 # Delete heavy data (it will be computed again if required)
372 # (it is required only when MSE is wanted in self.predict)
373 if self.verbose:
374 print("Light storage mode specified. "
375 + "Flushing autocorrelation matrix...")
376 self.D = None
377 self.ij = None
378 self.F = None
379 self.C = None
380 self.Ft = None
381 self.G = None
382
383 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: predict at line 385
Total time: 0 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
385 def predict(self, X, eval_MSE=False, batch_size=None):
386 """
387 This function evaluates the Gaussian Process model at x.
388
389 Parameters
390 ----------
391 X : array_like
392 An array with shape (n_eval, n_features) giving the point(s) at
393 which the prediction(s) should be made.
394
395 eval_MSE : boolean, optional
396 A boolean specifying whether the Mean Squared Error should be
397 evaluated or not.
398 Default assumes evalMSE = False and evaluates only the BLUP (mean
399 prediction).
400
401 batch_size : integer, optional
402 An integer giving the maximum number of points that can be
403 evaluated simulatneously (depending on the available memory).
404 Default is None so that all given points are evaluated at the same
405 time.
406
407 Returns
408 -------
409 y : array_like
410 An array with shape (n_eval, ) with the Best Linear Unbiased
411 Prediction at x.
412
413 MSE : array_like, optional (if eval_MSE == True)
414 An array with shape (n_eval, ) with the Mean Squared Error at x.
415 """
416
417 # Check input shapes
418 X = array2d(X)
419 n_eval, n_features_X = X.shape
420 n_samples, n_features = self.X.shape
421
422 # Run input checks
423 self._check_params(n_samples)
424
425 if n_features_X != n_features:
426 raise ValueError(("The number of features in X (X.shape[1] = %d) "
427 + "should match the sample size used for fit() "
428 + "which is %d.") % (n_features_X, n_features))
429
430 if batch_size is None:
431 # No memory management
432 # (evaluates all given points in a single batch run)
433
434 # Normalize input
435 X = (X - self.X_mean) / self.X_std
436
437 # Initialize output
438 y = np.zeros(n_eval)
439 if eval_MSE:
440 MSE = np.zeros(n_eval)
441
442 # Get pairwise componentwise L1-distances to the input training set
443 dx = manhattan_distances(X, Y=self.X, sum_over_features=False)
444 # Get regression function and correlation
445 f = self.regr(X)
446 r = self.corr(self.theta_, dx).reshape(n_eval, n_samples)
447
448 # Scaled predictor
449 y_ = np.dot(f, self.beta) + np.dot(r, self.gamma)
450
451 # Predictor
452 y = (self.y_mean + self.y_std * y_).ravel()
453
454 # Mean Squared Error
455 if eval_MSE:
456 C = self.C
457 if C is None:
458 # Light storage mode (need to recompute C, F, Ft and G)
459 if self.verbose:
460 print("This GaussianProcess used 'light' storage mode "
461 + "at instanciation. Need to recompute "
462 + "autocorrelation matrix...")
463 reduced_likelihood_function_value, par = \
464 self.reduced_likelihood_function()
465 self.C = par['C']
466 self.Ft = par['Ft']
467 self.G = par['G']
468
469 rt = solve_triangular(self.C, r.T, lower=True)
470
471 if self.beta0 is None:
472 # Universal Kriging
473 u = solve_triangular(self.G.T,
474 np.dot(self.Ft.T, rt) - f.T)
475 else:
476 # Ordinary Kriging
477 u = np.zeros(y.shape)
478
479 MSE = self.sigma2 * (1. - (rt ** 2.).sum(axis=0)
480 + (u ** 2.).sum(axis=0))
481
482 # Mean Squared Error might be slightly negative depending on
483 # machine precision: force to zero!
484 MSE[MSE < 0.] = 0.
485
486 return y, MSE
487
488 else:
489
490 return y
491
492 else:
493 # Memory management
494
495 if type(batch_size) is not int or batch_size <= 0:
496 raise Exception("batch_size must be a positive integer")
497
498 if eval_MSE:
499
500 y, MSE = np.zeros(n_eval), np.zeros(n_eval)
501 for k in range(max(1, n_eval / batch_size)):
502 batch_from = k * batch_size
503 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
504 y[batch_from:batch_to], MSE[batch_from:batch_to] = \
505 self.predict(X[batch_from:batch_to],
506 eval_MSE=eval_MSE, batch_size=None)
507
508 return y, MSE
509
510 else:
511
512 y = np.zeros(n_eval)
513 for k in range(max(1, n_eval / batch_size)):
514 batch_from = k * batch_size
515 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
516 y[batch_from:batch_to] = \
517 self.predict(X[batch_from:batch_to],
518 eval_MSE=eval_MSE, batch_size=None)
519
520 return y
Benchmark statement
obj.predict(X_t)
Execution time
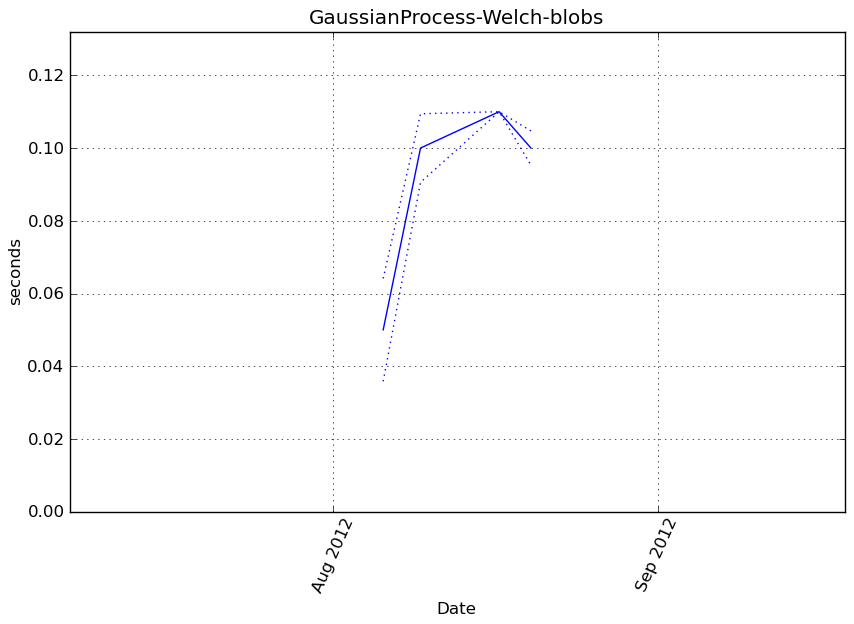
Memory usage
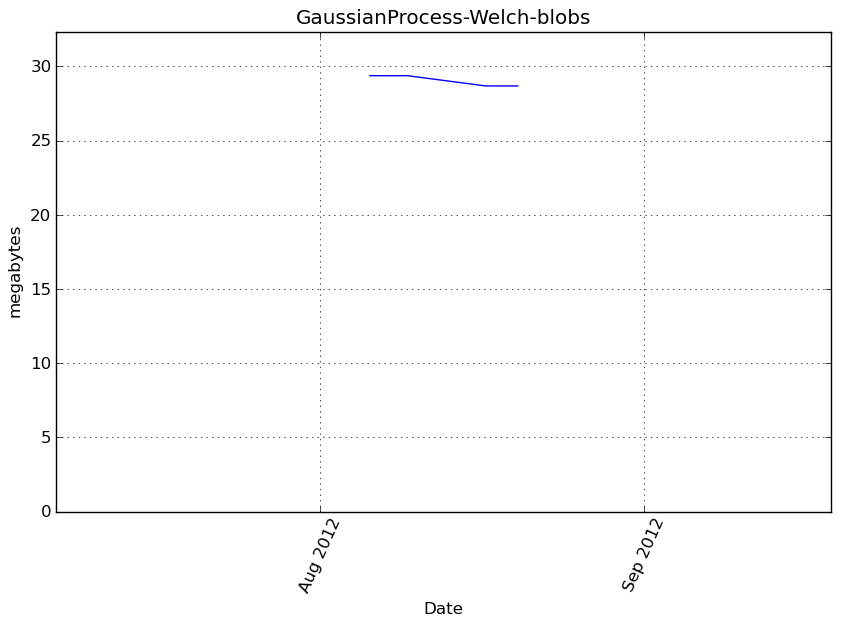
Additional output
cProfile
133 function calls in 0.099 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.099 0.099 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.099 0.099 <f>:1(<module>)
1 0.000 0.000 0.099 0.099 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:385(predict)
1 0.074 0.074 0.075 0.075 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:188(manhattan_distances)
1 0.014 0.014 0.024 0.024 /tmp/vb_sklearn/sklearn/gaussian_process/correlation_models.py:58(squared_exponential)
5 0.010 0.002 0.010 0.002 {method 'sum' of 'numpy.ndarray' objects}
1 0.000 0.000 0.010 0.010 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/metrics/pairwise.py:52(check_pairwise_arrays)
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:10(assert_all_finite)
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:70(atleast2d_or_csr)
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:23(safe_asarray)
4 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:62(array2d)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py:822(_check_params)
8 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/base.py:553(isspmatrix)
2 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
8 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/sparse/sputils.py:116(_isinstance)
4 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/shape_base.py:58(atleast_2d)
1 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/gaussian_process/regression_models.py:16(constant)
10 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
14 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1508(any)
2 0.000 0.000 0.000 0.000 {method 'any' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:237(asanyarray)
9 0.000 0.000 0.000 0.000 {isinstance}
16 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
2 0.000 0.000 0.000 0.000 {method 'reshape' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
14 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
2 0.000 0.000 0.000 0.000 {callable}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: fit at line 250
Total time: 0.143732 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
250 def fit(self, X, y):
251 """
252 The Gaussian Process model fitting method.
253
254 Parameters
255 ----------
256 X : double array_like
257 An array with shape (n_samples, n_features) with the input at which
258 observations were made.
259
260 y : double array_like
261 An array with shape (n_features, ) with the observations of the
262 scalar output to be predicted.
263
264 Returns
265 -------
266 gp : self
267 A fitted Gaussian Process model object awaiting data to perform
268 predictions.
269 """
270 1 13 13.0 0.0 self.random_state = check_random_state(self.random_state)
271
272 # Force data to 2D numpy.array
273 1 61 61.0 0.0 X = array2d(X)
274 1 20 20.0 0.0 y = np.asarray(y).ravel()[:, np.newaxis]
275
276 # Check shapes of DOE & observations
277 1 5 5.0 0.0 n_samples_X, n_features = X.shape
278 1 5 5.0 0.0 n_samples_y = y.shape[0]
279
280 1 4 4.0 0.0 if n_samples_X != n_samples_y:
281 raise ValueError("X and y must have the same number of rows.")
282 else:
283 1 4 4.0 0.0 n_samples = n_samples_X
284
285 # Run input checks
286 1 188 188.0 0.1 self._check_params(n_samples)
287
288 # Normalize data or don't
289 1 5 5.0 0.0 if self.normalize:
290 1 89 89.0 0.1 X_mean = np.mean(X, axis=0)
291 1 280 280.0 0.2 X_std = np.std(X, axis=0)
292 1 44 44.0 0.0 y_mean = np.mean(y, axis=0)
293 1 85 85.0 0.1 y_std = np.std(y, axis=0)
294 1 23 23.0 0.0 X_std[X_std == 0.] = 1.
295 1 17 17.0 0.0 y_std[y_std == 0.] = 1.
296 # center and scale X if necessary
297 1 404 404.0 0.3 X = (X - X_mean) / X_std
298 1 42 42.0 0.0 y = (y - y_mean) / y_std
299 else:
300 X_mean = np.zeros(1)
301 X_std = np.ones(1)
302 y_mean = np.zeros(1)
303 y_std = np.ones(1)
304
305 # Calculate matrix of distances D between samples
306 1 74826 74826.0 52.1 D, ij = l1_cross_distances(X)
307 1 7645 7645.0 5.3 if np.min(np.sum(D, axis=1)) == 0. \
308 and self.corr != correlation.pure_nugget:
309 raise Exception("Multiple input features cannot have the same"
310 " value")
311
312 # Regression matrix and parameters
313 1 51 51.0 0.0 F = self.regr(X)
314 1 5 5.0 0.0 n_samples_F = F.shape[0]
315 1 6 6.0 0.0 if F.ndim > 1:
316 1 5 5.0 0.0 p = F.shape[1]
317 else:
318 p = 1
319 1 5 5.0 0.0 if n_samples_F != n_samples:
320 raise Exception("Number of rows in F and X do not match. Most "
321 + "likely something is going wrong with the "
322 + "regression model.")
323 1 4 4.0 0.0 if p > n_samples_F:
324 raise Exception(("Ordinary least squares problem is undetermined "
325 + "n_samples=%d must be greater than the "
326 + "regression model size p=%d.") % (n_samples, p))
327 1 5 5.0 0.0 if self.beta0 is not None:
328 if self.beta0.shape[0] != p:
329 raise Exception("Shapes of beta0 and F do not match.")
330
331 # Set attributes
332 1 6 6.0 0.0 self.X = X
333 1 4 4.0 0.0 self.y = y
334 1 6 6.0 0.0 self.D = D
335 1 4 4.0 0.0 self.ij = ij
336 1 5 5.0 0.0 self.F = F
337 1 5 5.0 0.0 self.X_mean, self.X_std = X_mean, X_std
338 1 7 7.0 0.0 self.y_mean, self.y_std = y_mean, y_std
339
340 # Determine Gaussian Process model parameters
341 1 6 6.0 0.0 if self.thetaL is not None and self.thetaU is not None:
342 # Maximum Likelihood Estimation of the parameters
343 if self.verbose:
344 print("Performing Maximum Likelihood Estimation of the "
345 + "autocorrelation parameters...")
346 self.theta_, self.reduced_likelihood_function_value_, par = \
347 self._arg_max_reduced_likelihood_function()
348 if np.isinf(self.reduced_likelihood_function_value_):
349 raise Exception("Bad parameter region. "
350 + "Try increasing upper bound")
351
352 else:
353 # Given parameters
354 1 4 4.0 0.0 if self.verbose:
355 print("Given autocorrelation parameters. "
356 + "Computing Gaussian Process model parameters...")
357 1 5 5.0 0.0 self.theta_ = self.theta0
358 self.reduced_likelihood_function_value_, par = \
359 1 59780 59780.0 41.6 self.reduced_likelihood_function()
360 1 21 21.0 0.0 if np.isinf(self.reduced_likelihood_function_value_):
361 raise Exception("Bad point. Try increasing theta0.")
362
363 1 5 5.0 0.0 self.beta = par['beta']
364 1 5 5.0 0.0 self.gamma = par['gamma']
365 1 5 5.0 0.0 self.sigma2 = par['sigma2']
366 1 5 5.0 0.0 self.C = par['C']
367 1 5 5.0 0.0 self.Ft = par['Ft']
368 1 4 4.0 0.0 self.G = par['G']
369
370 1 5 5.0 0.0 if self.storage_mode == 'light':
371 # Delete heavy data (it will be computed again if required)
372 # (it is required only when MSE is wanted in self.predict)
373 if self.verbose:
374 print("Light storage mode specified. "
375 + "Flushing autocorrelation matrix...")
376 self.D = None
377 self.ij = None
378 self.F = None
379 self.C = None
380 self.Ft = None
381 self.G = None
382
383 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/gaussian_process/gaussian_process.py
Function: predict at line 385
Total time: 0.099102 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
385 def predict(self, X, eval_MSE=False, batch_size=None):
386 """
387 This function evaluates the Gaussian Process model at x.
388
389 Parameters
390 ----------
391 X : array_like
392 An array with shape (n_eval, n_features) giving the point(s) at
393 which the prediction(s) should be made.
394
395 eval_MSE : boolean, optional
396 A boolean specifying whether the Mean Squared Error should be
397 evaluated or not.
398 Default assumes evalMSE = False and evaluates only the BLUP (mean
399 prediction).
400
401 batch_size : integer, optional
402 An integer giving the maximum number of points that can be
403 evaluated simulatneously (depending on the available memory).
404 Default is None so that all given points are evaluated at the same
405 time.
406
407 Returns
408 -------
409 y : array_like
410 An array with shape (n_eval, ) with the Best Linear Unbiased
411 Prediction at x.
412
413 MSE : array_like, optional (if eval_MSE == True)
414 An array with shape (n_eval, ) with the Mean Squared Error at x.
415 """
416
417 # Check input shapes
418 1 69 69.0 0.1 X = array2d(X)
419 1 5 5.0 0.0 n_eval, n_features_X = X.shape
420 1 5 5.0 0.0 n_samples, n_features = self.X.shape
421
422 # Run input checks
423 1 185 185.0 0.2 self._check_params(n_samples)
424
425 1 5 5.0 0.0 if n_features_X != n_features:
426 raise ValueError(("The number of features in X (X.shape[1] = %d) "
427 + "should match the sample size used for fit() "
428 + "which is %d.") % (n_features_X, n_features))
429
430 1 3 3.0 0.0 if batch_size is None:
431 # No memory management
432 # (evaluates all given points in a single batch run)
433
434 # Normalize input
435 1 289 289.0 0.3 X = (X - self.X_mean) / self.X_std
436
437 # Initialize output
438 1 15 15.0 0.0 y = np.zeros(n_eval)
439 1 4 4.0 0.0 if eval_MSE:
440 MSE = np.zeros(n_eval)
441
442 # Get pairwise componentwise L1-distances to the input training set
443 1 74477 74477.0 75.2 dx = manhattan_distances(X, Y=self.X, sum_over_features=False)
444 # Get regression function and correlation
445 1 58 58.0 0.1 f = self.regr(X)
446 1 23794 23794.0 24.0 r = self.corr(self.theta_, dx).reshape(n_eval, n_samples)
447
448 # Scaled predictor
449 1 145 145.0 0.1 y_ = np.dot(f, self.beta) + np.dot(r, self.gamma)
450
451 # Predictor
452 1 40 40.0 0.0 y = (self.y_mean + self.y_std * y_).ravel()
453
454 # Mean Squared Error
455 1 4 4.0 0.0 if eval_MSE:
456 C = self.C
457 if C is None:
458 # Light storage mode (need to recompute C, F, Ft and G)
459 if self.verbose:
460 print("This GaussianProcess used 'light' storage mode "
461 + "at instanciation. Need to recompute "
462 + "autocorrelation matrix...")
463 reduced_likelihood_function_value, par = \
464 self.reduced_likelihood_function()
465 self.C = par['C']
466 self.Ft = par['Ft']
467 self.G = par['G']
468
469 rt = solve_triangular(self.C, r.T, lower=True)
470
471 if self.beta0 is None:
472 # Universal Kriging
473 u = solve_triangular(self.G.T,
474 np.dot(self.Ft.T, rt) - f.T)
475 else:
476 # Ordinary Kriging
477 u = np.zeros(y.shape)
478
479 MSE = self.sigma2 * (1. - (rt ** 2.).sum(axis=0)
480 + (u ** 2.).sum(axis=0))
481
482 # Mean Squared Error might be slightly negative depending on
483 # machine precision: force to zero!
484 MSE[MSE < 0.] = 0.
485
486 return y, MSE
487
488 else:
489
490 1 4 4.0 0.0 return y
491
492 else:
493 # Memory management
494
495 if type(batch_size) is not int or batch_size <= 0:
496 raise Exception("batch_size must be a positive integer")
497
498 if eval_MSE:
499
500 y, MSE = np.zeros(n_eval), np.zeros(n_eval)
501 for k in range(max(1, n_eval / batch_size)):
502 batch_from = k * batch_size
503 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
504 y[batch_from:batch_to], MSE[batch_from:batch_to] = \
505 self.predict(X[batch_from:batch_to],
506 eval_MSE=eval_MSE, batch_size=None)
507
508 return y, MSE
509
510 else:
511
512 y = np.zeros(n_eval)
513 for k in range(max(1, n_eval / batch_size)):
514 batch_from = k * batch_size
515 batch_to = min([(k + 1) * batch_size + 1, n_eval + 1])
516 y[batch_from:batch_to] = \
517 self.predict(X[batch_from:batch_to],
518 eval_MSE=eval_MSE, batch_size=None)
519
520 return y
