Benchmarks for pls¶
CCA-minimadelon¶
Benchmark setup
from sklearn.pls import CCA from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('minimadelon') obj = CCA(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
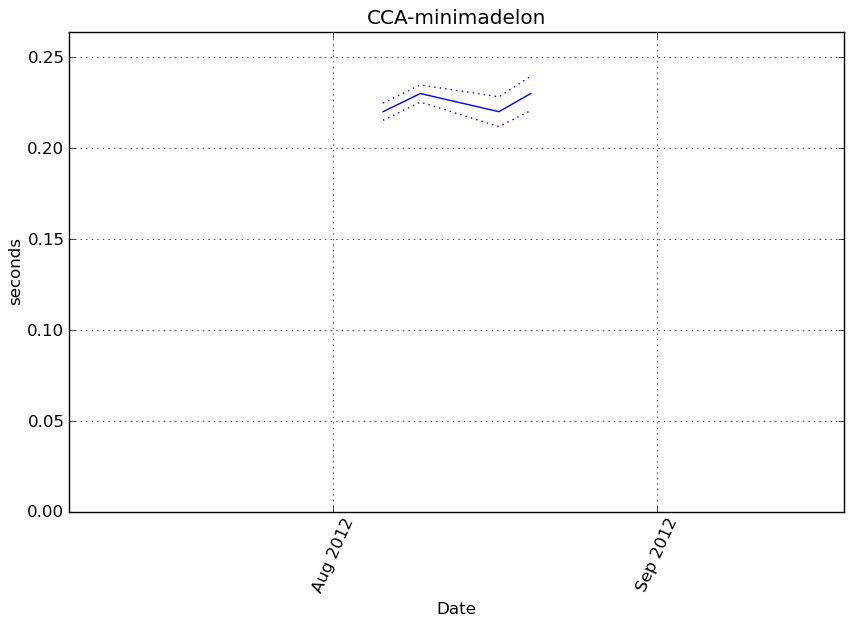
Memory usage
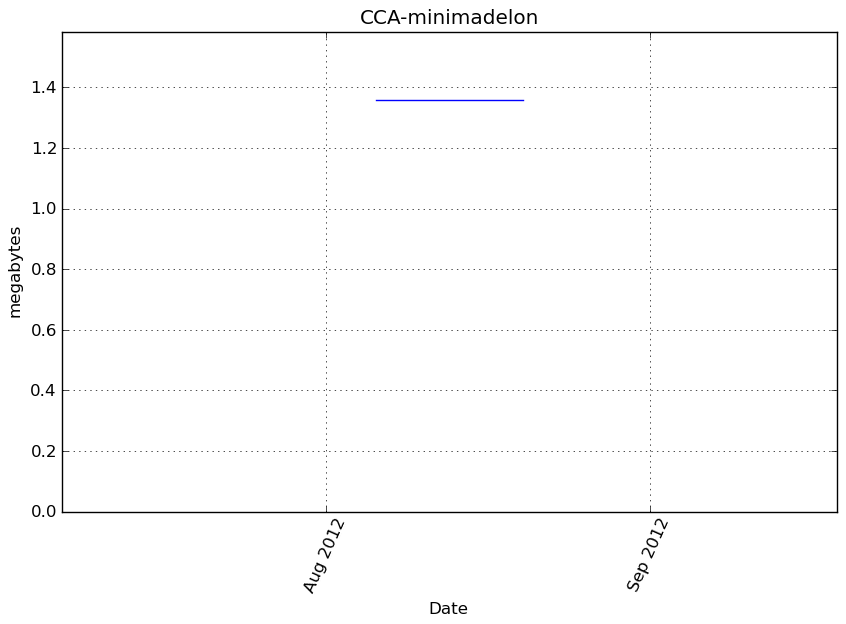
Additional output
cProfile
8312 function calls in 0.205 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.205 0.205 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.205 0.205 <f>:1(<module>)
1 0.001 0.001 0.205 0.205 /tmp/vb_sklearn/sklearn/pls.py:218(fit)
2 0.089 0.044 0.201 0.101 /tmp/vb_sklearn/sklearn/pls.py:16(_nipals_twoblocks_inner_loop)
8023 0.094 0.000 0.094 0.000 {numpy.core._dotblas.dot}
4 0.000 0.000 0.020 0.005 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:457(pinv)
4 0.016 0.004 0.018 0.005 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:365(lstsq)
14 0.002 0.000 0.003 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py:526(asarray_chkfinite)
4 0.000 0.000 0.001 0.000 {map}
1 0.001 0.001 0.001 0.001 /tmp/vb_sklearn/sklearn/pls.py:75(_center_scale_xy)
2 0.001 0.000 0.001 0.000 {method 'std' of 'numpy.ndarray' objects}
28 0.001 0.000 0.001 0.000 {method 'any' of 'numpy.ndarray' objects}
6 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:60(get_lapack_funcs)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:253(inv)
6 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:45(find_best_lapack_type)
18 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
2 0.000 0.000 0.000 0.000 {method 'mean' of 'numpy.ndarray' objects}
2 0.000 0.000 0.000 0.000 /tmp/vb_sklearn/sklearn/utils/validation.py:33(as_float_array)
18 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
2 0.000 0.000 0.000 0.000 {method 'copy' of 'numpy.ndarray' objects}
12 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
4 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1830(identity)
10 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:23(cast_to_lapack_prefix)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/getlimits.py:91(__new__)
2 0.000 0.000 0.000 0.000 {_warnings.warn}
16 0.000 0.000 0.000 0.000 {getattr}
4 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.generic' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/fromnumeric.py:1379(sum)
2 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
1 0.000 0.000 0.000 0.000 {method 'sum' of 'numpy.ndarray' objects}
20 0.000 0.000 0.000 0.000 {issubclass}
12 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
12 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
7 0.000 0.000 0.000 0.000 {isinstance}
6 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:449(isfortran)
6 0.000 0.000 0.000 0.000 {method 'sort' of 'list' objects}
10 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/misc.py:22(_datacopied)
6 0.000 0.000 0.000 0.000 {range}
18 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
18 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {method 'reshape' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/pls.py
Function: fit at line 218
Total time: 0.223438 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
218 def fit(self, X, Y):
219 # copy since this will contains the residuals (deflated) matrices
220 1 53 53.0 0.0 X = as_float_array(X, copy=self.copy)
221 1 20 20.0 0.0 Y = as_float_array(Y, copy=self.copy)
222
223 1 4 4.0 0.0 if X.ndim != 2:
224 raise ValueError('X must be a 2D array')
225 1 4 4.0 0.0 if Y.ndim == 1:
226 Y = Y.reshape((Y.size, 1))
227 1 4 4.0 0.0 if Y.ndim != 2:
228 raise ValueError('Y must be a 1D or a 2D array')
229
230 1 5 5.0 0.0 n = X.shape[0]
231 1 5 5.0 0.0 p = X.shape[1]
232 1 4 4.0 0.0 q = Y.shape[1]
233
234 1 5 5.0 0.0 if n != Y.shape[0]:
235 raise ValueError(
236 'Incompatible shapes: X has %s samples, while Y '
237 'has %s' % (X.shape[0], Y.shape[0]))
238 1 5 5.0 0.0 if self.n_components < 1 or self.n_components > p:
239 raise ValueError('invalid number of components')
240 1 4 4.0 0.0 if self.algorithm not in ("svd", "nipals"):
241 raise ValueError("Got algorithm %s when only 'svd' "
242 "and 'nipals' are known" % self.algorithm)
243 1 4 4.0 0.0 if self.algorithm == "svd" and self.mode == "B":
244 raise ValueError('Incompatible configuration: mode B is not '
245 'implemented with svd algorithm')
246 1 4 4.0 0.0 if not self.deflation_mode in ["canonical", "regression"]:
247 raise ValueError('The deflation mode is unknown')
248 # Scale (in place)
249 X, Y, self.x_mean_, self.y_mean_, self.x_std_, self.y_std_\
250 1 1020 1020.0 0.5 = _center_scale_xy(X, Y, self.scale)
251 # Residuals (deflated) matrices
252 1 15 15.0 0.0 Xk = X
253 1 4 4.0 0.0 Yk = Y
254 # Results matrices
255 1 11 11.0 0.0 self.x_scores_ = np.zeros((n, self.n_components))
256 1 6 6.0 0.0 self.y_scores_ = np.zeros((n, self.n_components))
257 1 9 9.0 0.0 self.x_weights_ = np.zeros((p, self.n_components))
258 1 6 6.0 0.0 self.y_weights_ = np.zeros((q, self.n_components))
259 1 8 8.0 0.0 self.x_loadings_ = np.zeros((p, self.n_components))
260 1 6 6.0 0.0 self.y_loadings_ = np.zeros((q, self.n_components))
261
262 # NIPALS algo: outer loop, over components
263 3 18 6.0 0.0 for k in xrange(self.n_components):
264 #1) weights estimation (inner loop)
265 # -----------------------------------
266 2 9 4.5 0.0 if self.algorithm == "nipals":
267 2 7 3.5 0.0 x_weights, y_weights = _nipals_twoblocks_inner_loop(
268 2 8 4.0 0.0 X=Xk, Y=Yk, mode=self.mode,
269 2 10 5.0 0.0 max_iter=self.max_iter, tol=self.tol,
270 2 219636 109818.0 98.3 norm_y_weights=self.norm_y_weights)
271 elif self.algorithm == "svd":
272 x_weights, y_weights = _svd_cross_product(X=Xk, Y=Yk)
273 # compute scores
274 2 110 55.0 0.0 x_scores = np.dot(Xk, x_weights)
275 2 9 4.5 0.0 if self.norm_y_weights:
276 2 8 4.0 0.0 y_ss = 1
277 else:
278 y_ss = np.dot(y_weights.T, y_weights)
279 2 60 30.0 0.0 y_scores = np.dot(Yk, y_weights) / y_ss
280 # test for null variance
281 2 109 54.5 0.0 if np.dot(x_scores.T, x_scores) < np.finfo(np.double).eps:
282 warnings.warn('X scores are null at iteration %s' % k)
283 #2) Deflation (in place)
284 # ----------------------
285 # Possible memory footprint reduction may done here: in order to
286 # avoid the allocation of a data chunk for the rank-one
287 # approximations matrix which is then substracted to Xk, we suggest
288 # to perform a column-wise deflation.
289 #
290 # - regress Xk's on x_score
291 2 154 77.0 0.1 x_loadings = np.dot(Xk.T, x_scores) / np.dot(x_scores.T, x_scores)
292 # - substract rank-one approximations to obtain remainder matrix
293 2 533 266.5 0.2 Xk -= np.dot(x_scores, x_loadings.T)
294 2 13 6.5 0.0 if self.deflation_mode == "canonical":
295 # - regress Yk's on y_score, then substract rank-one approx.
296 2 33 16.5 0.0 y_loadings = np.dot(Yk.T, y_scores) \
297 2 55 27.5 0.0 / np.dot(y_scores.T, y_scores)
298 2 75 37.5 0.0 Yk -= np.dot(y_scores, y_loadings.T)
299 2 10 5.0 0.0 if self.deflation_mode == "regression":
300 # - regress Yk's on x_score, then substract rank-one approx.
301 y_loadings = np.dot(Yk.T, x_scores) \
302 / np.dot(x_scores.T, x_scores)
303 Yk -= np.dot(x_scores, y_loadings.T)
304 # 3) Store weights, scores and loadings # Notation:
305 2 48 24.0 0.0 self.x_scores_[:, k] = x_scores.ravel() # T
306 2 24 12.0 0.0 self.y_scores_[:, k] = y_scores.ravel() # U
307 2 25 12.5 0.0 self.x_weights_[:, k] = x_weights.ravel() # W
308 2 22 11.0 0.0 self.y_weights_[:, k] = y_weights.ravel() # C
309 2 25 12.5 0.0 self.x_loadings_[:, k] = x_loadings.ravel() # P
310 2 22 11.0 0.0 self.y_loadings_[:, k] = y_loadings.ravel() # Q
311 # Such that: X = TP' + Err and Y = UQ' + Err
312
313 # 4) rotations from input space to transformed space (scores)
314 # T = X W(P'W)^-1 = XW* (W* : p x k matrix)
315 # U = Y C(Q'C)^-1 = YC* (W* : q x k matrix)
316 1 5 5.0 0.0 self.x_rotations_ = np.dot(self.x_weights_,
317 1 617 617.0 0.3 linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
318 1 7 7.0 0.0 if Y.shape[1] > 1:
319 1 5 5.0 0.0 self.y_rotations_ = np.dot(self.y_weights_,
320 1 263 263.0 0.1 linalg.inv(np.dot(self.y_loadings_.T, self.y_weights_)))
321 else:
322 self.y_rotations_ = np.ones(1)
323
324 1 4 4.0 0.0 if True or self.deflation_mode == "regression":
325 # Estimate regression coefficient
326 # Regress Y on T
327 # Y = TQ' + Err,
328 # Then express in function of X
329 # Y = X W(P'W)^-1Q' + Err = XB + Err
330 # => B = W*Q' (p x q)
331 1 168 168.0 0.1 self.coefs = np.dot(self.x_rotations_, self.y_loadings_.T)
332 self.coefs = 1. / self.x_std_.reshape((p, 1)) * \
333 1 136 136.0 0.1 self.coefs * self.y_std_
334 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/pls.py
Function: transform at line 336
Total time: 0 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
336 def transform(self, X, Y=None, copy=True):
337 """Apply the dimension reduction learned on the train data.
338
339 Parameters
340 ----------
341 X : array-like of predictors, shape = [n_samples, p]
342 Training vectors, where n_samples in the number of samples and
343 p is the number of predictors.
344
345 Y : array-like of response, shape = [n_samples, q], optional
346 Training vectors, where n_samples in the number of samples and
347 q is the number of response variables.
348
349 copy : boolean
350 Whether to copy X and Y, or perform in-place normalization.
351
352 Returns
353 -------
354 x_scores if Y is not given, (x_scores, y_scores) otherwise.
355 """
356 # Normalize
357 if copy:
358 Xc = (np.asarray(X) - self.x_mean_) / self.x_std_
359 if Y is not None:
360 Yc = (np.asarray(Y) - self.y_mean_) / self.y_std_
361 else:
362 X = np.asarray(X)
363 Xc -= self.x_mean_
364 Xc /= self.x_std_
365 if Y is not None:
366 Y = np.asarray(Y)
367 Yc -= self.y_mean_
368 Yc /= self.y_std_
369 # Apply rotation
370 x_scores = np.dot(Xc, self.x_rotations_)
371 if Y is not None:
372 y_scores = np.dot(Yc, self.y_rotations_)
373 return x_scores, y_scores
374
375 return x_scores
Benchmark statement
obj.transform(X)
Execution time
Memory usage
Additional output
cProfile
7 function calls in 0.001 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.001 0.001 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.001 0.001 <f>:1(<module>)
1 0.000 0.000 0.001 0.001 /tmp/vb_sklearn/sklearn/pls.py:336(transform)
1 0.000 0.000 0.000 0.000 {numpy.core._dotblas.dot}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/pls.py
Function: fit at line 218
Total time: 0.223438 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
218 def fit(self, X, Y):
219 # copy since this will contains the residuals (deflated) matrices
220 1 53 53.0 0.0 X = as_float_array(X, copy=self.copy)
221 1 20 20.0 0.0 Y = as_float_array(Y, copy=self.copy)
222
223 1 4 4.0 0.0 if X.ndim != 2:
224 raise ValueError('X must be a 2D array')
225 1 4 4.0 0.0 if Y.ndim == 1:
226 Y = Y.reshape((Y.size, 1))
227 1 4 4.0 0.0 if Y.ndim != 2:
228 raise ValueError('Y must be a 1D or a 2D array')
229
230 1 5 5.0 0.0 n = X.shape[0]
231 1 5 5.0 0.0 p = X.shape[1]
232 1 4 4.0 0.0 q = Y.shape[1]
233
234 1 5 5.0 0.0 if n != Y.shape[0]:
235 raise ValueError(
236 'Incompatible shapes: X has %s samples, while Y '
237 'has %s' % (X.shape[0], Y.shape[0]))
238 1 5 5.0 0.0 if self.n_components < 1 or self.n_components > p:
239 raise ValueError('invalid number of components')
240 1 4 4.0 0.0 if self.algorithm not in ("svd", "nipals"):
241 raise ValueError("Got algorithm %s when only 'svd' "
242 "and 'nipals' are known" % self.algorithm)
243 1 4 4.0 0.0 if self.algorithm == "svd" and self.mode == "B":
244 raise ValueError('Incompatible configuration: mode B is not '
245 'implemented with svd algorithm')
246 1 4 4.0 0.0 if not self.deflation_mode in ["canonical", "regression"]:
247 raise ValueError('The deflation mode is unknown')
248 # Scale (in place)
249 X, Y, self.x_mean_, self.y_mean_, self.x_std_, self.y_std_\
250 1 1020 1020.0 0.5 = _center_scale_xy(X, Y, self.scale)
251 # Residuals (deflated) matrices
252 1 15 15.0 0.0 Xk = X
253 1 4 4.0 0.0 Yk = Y
254 # Results matrices
255 1 11 11.0 0.0 self.x_scores_ = np.zeros((n, self.n_components))
256 1 6 6.0 0.0 self.y_scores_ = np.zeros((n, self.n_components))
257 1 9 9.0 0.0 self.x_weights_ = np.zeros((p, self.n_components))
258 1 6 6.0 0.0 self.y_weights_ = np.zeros((q, self.n_components))
259 1 8 8.0 0.0 self.x_loadings_ = np.zeros((p, self.n_components))
260 1 6 6.0 0.0 self.y_loadings_ = np.zeros((q, self.n_components))
261
262 # NIPALS algo: outer loop, over components
263 3 18 6.0 0.0 for k in xrange(self.n_components):
264 #1) weights estimation (inner loop)
265 # -----------------------------------
266 2 9 4.5 0.0 if self.algorithm == "nipals":
267 2 7 3.5 0.0 x_weights, y_weights = _nipals_twoblocks_inner_loop(
268 2 8 4.0 0.0 X=Xk, Y=Yk, mode=self.mode,
269 2 10 5.0 0.0 max_iter=self.max_iter, tol=self.tol,
270 2 219636 109818.0 98.3 norm_y_weights=self.norm_y_weights)
271 elif self.algorithm == "svd":
272 x_weights, y_weights = _svd_cross_product(X=Xk, Y=Yk)
273 # compute scores
274 2 110 55.0 0.0 x_scores = np.dot(Xk, x_weights)
275 2 9 4.5 0.0 if self.norm_y_weights:
276 2 8 4.0 0.0 y_ss = 1
277 else:
278 y_ss = np.dot(y_weights.T, y_weights)
279 2 60 30.0 0.0 y_scores = np.dot(Yk, y_weights) / y_ss
280 # test for null variance
281 2 109 54.5 0.0 if np.dot(x_scores.T, x_scores) < np.finfo(np.double).eps:
282 warnings.warn('X scores are null at iteration %s' % k)
283 #2) Deflation (in place)
284 # ----------------------
285 # Possible memory footprint reduction may done here: in order to
286 # avoid the allocation of a data chunk for the rank-one
287 # approximations matrix which is then substracted to Xk, we suggest
288 # to perform a column-wise deflation.
289 #
290 # - regress Xk's on x_score
291 2 154 77.0 0.1 x_loadings = np.dot(Xk.T, x_scores) / np.dot(x_scores.T, x_scores)
292 # - substract rank-one approximations to obtain remainder matrix
293 2 533 266.5 0.2 Xk -= np.dot(x_scores, x_loadings.T)
294 2 13 6.5 0.0 if self.deflation_mode == "canonical":
295 # - regress Yk's on y_score, then substract rank-one approx.
296 2 33 16.5 0.0 y_loadings = np.dot(Yk.T, y_scores) \
297 2 55 27.5 0.0 / np.dot(y_scores.T, y_scores)
298 2 75 37.5 0.0 Yk -= np.dot(y_scores, y_loadings.T)
299 2 10 5.0 0.0 if self.deflation_mode == "regression":
300 # - regress Yk's on x_score, then substract rank-one approx.
301 y_loadings = np.dot(Yk.T, x_scores) \
302 / np.dot(x_scores.T, x_scores)
303 Yk -= np.dot(x_scores, y_loadings.T)
304 # 3) Store weights, scores and loadings # Notation:
305 2 48 24.0 0.0 self.x_scores_[:, k] = x_scores.ravel() # T
306 2 24 12.0 0.0 self.y_scores_[:, k] = y_scores.ravel() # U
307 2 25 12.5 0.0 self.x_weights_[:, k] = x_weights.ravel() # W
308 2 22 11.0 0.0 self.y_weights_[:, k] = y_weights.ravel() # C
309 2 25 12.5 0.0 self.x_loadings_[:, k] = x_loadings.ravel() # P
310 2 22 11.0 0.0 self.y_loadings_[:, k] = y_loadings.ravel() # Q
311 # Such that: X = TP' + Err and Y = UQ' + Err
312
313 # 4) rotations from input space to transformed space (scores)
314 # T = X W(P'W)^-1 = XW* (W* : p x k matrix)
315 # U = Y C(Q'C)^-1 = YC* (W* : q x k matrix)
316 1 5 5.0 0.0 self.x_rotations_ = np.dot(self.x_weights_,
317 1 617 617.0 0.3 linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
318 1 7 7.0 0.0 if Y.shape[1] > 1:
319 1 5 5.0 0.0 self.y_rotations_ = np.dot(self.y_weights_,
320 1 263 263.0 0.1 linalg.inv(np.dot(self.y_loadings_.T, self.y_weights_)))
321 else:
322 self.y_rotations_ = np.ones(1)
323
324 1 4 4.0 0.0 if True or self.deflation_mode == "regression":
325 # Estimate regression coefficient
326 # Regress Y on T
327 # Y = TQ' + Err,
328 # Then express in function of X
329 # Y = X W(P'W)^-1Q' + Err = XB + Err
330 # => B = W*Q' (p x q)
331 1 168 168.0 0.1 self.coefs = np.dot(self.x_rotations_, self.y_loadings_.T)
332 self.coefs = 1. / self.x_std_.reshape((p, 1)) * \
333 1 136 136.0 0.1 self.coefs * self.y_std_
334 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/pls.py
Function: transform at line 336
Total time: 0.000648 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
336 def transform(self, X, Y=None, copy=True):
337 """Apply the dimension reduction learned on the train data.
338
339 Parameters
340 ----------
341 X : array-like of predictors, shape = [n_samples, p]
342 Training vectors, where n_samples in the number of samples and
343 p is the number of predictors.
344
345 Y : array-like of response, shape = [n_samples, q], optional
346 Training vectors, where n_samples in the number of samples and
347 q is the number of response variables.
348
349 copy : boolean
350 Whether to copy X and Y, or perform in-place normalization.
351
352 Returns
353 -------
354 x_scores if Y is not given, (x_scores, y_scores) otherwise.
355 """
356 # Normalize
357 1 3 3.0 0.5 if copy:
358 1 413 413.0 63.7 Xc = (np.asarray(X) - self.x_mean_) / self.x_std_
359 1 2 2.0 0.3 if Y is not None:
360 Yc = (np.asarray(Y) - self.y_mean_) / self.y_std_
361 else:
362 X = np.asarray(X)
363 Xc -= self.x_mean_
364 Xc /= self.x_std_
365 if Y is not None:
366 Y = np.asarray(Y)
367 Yc -= self.y_mean_
368 Yc /= self.y_std_
369 # Apply rotation
370 1 225 225.0 34.7 x_scores = np.dot(Xc, self.x_rotations_)
371 1 3 3.0 0.5 if Y is not None:
372 y_scores = np.dot(Yc, self.y_rotations_)
373 return x_scores, y_scores
374
375 1 2 2.0 0.3 return x_scores
CCA-blobs¶
Benchmark setup
from sklearn.pls import CCA from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('blobs') obj = CCA(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
Memory usage
Additional output
Traceback
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 150, in run
exec step in ns
File "<string>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 260, in run
mem_usages = magic_memit(ns, self.code, repeat=self.mem_repeat)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 672, in magic_memit
raise RuntimeError('ERROR: all subprocesses exited unsuccessfully.'
RuntimeError: ERROR: all subprocesses exited unsuccessfully. Try again with the `-i` option.
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 293, in run
prof.runcall(f)
File "/sp/lib/python/cpython-2.7.2/lib/python2.7/cProfile.py", line 149, in runcall
return func(*args, **kw)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 288, in f
exec code in ns
File "<f>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 351, in run
prof.runcall(f)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/line_profiler.py", line 137, in runcall
return func(*args, **kw)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 348, in f
exec code in ns
File "<f>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Benchmark statement
obj.transform(X)
Execution time
Memory usage
Additional output
PLSCanonical-arcene¶
Benchmark setup
from sklearn.pls import PLSCanonical from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('arcene') obj = PLSCanonical(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
Memory usage
Additional output
Traceback
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 150, in run
exec step in ns
File "<string>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 260, in run
mem_usages = magic_memit(ns, self.code, repeat=self.mem_repeat)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 672, in magic_memit
raise RuntimeError('ERROR: all subprocesses exited unsuccessfully.'
RuntimeError: ERROR: all subprocesses exited unsuccessfully. Try again with the `-i` option.
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 293, in run
prof.runcall(f)
File "/sp/lib/python/cpython-2.7.2/lib/python2.7/cProfile.py", line 149, in runcall
return func(*args, **kw)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 288, in f
exec code in ns
File "<f>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 351, in run
prof.runcall(f)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/line_profiler.py", line 137, in runcall
return func(*args, **kw)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 348, in f
exec code in ns
File "<f>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Benchmark statement
obj.transform(X)
Execution time
Memory usage
Additional output
PLSCanonical-blobs¶
Benchmark setup
from sklearn.pls import PLSCanonical from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('blobs') obj = PLSCanonical(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
Memory usage
Additional output
Traceback
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 150, in run
exec step in ns
File "<string>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 260, in run
mem_usages = magic_memit(ns, self.code, repeat=self.mem_repeat)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 672, in magic_memit
raise RuntimeError('ERROR: all subprocesses exited unsuccessfully.'
RuntimeError: ERROR: all subprocesses exited unsuccessfully. Try again with the `-i` option.
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 293, in run
prof.runcall(f)
File "/sp/lib/python/cpython-2.7.2/lib/python2.7/cProfile.py", line 149, in runcall
return func(*args, **kw)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 288, in f
exec code in ns
File "<f>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Traceback (most recent call last):
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 351, in run
prof.runcall(f)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/line_profiler.py", line 137, in runcall
return func(*args, **kw)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py", line 348, in f
exec code in ns
File "<f>", line 1, in <module>
File "/tmp/vb_sklearn/sklearn/pls.py", line 317, in fit
linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py", line 287, in inv
a1 = np.asarray_chkfinite(a)
File "/home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py", line 590, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Benchmark statement
obj.transform(X)
Execution time
Memory usage
Additional output
PLSRegression-arcene¶
Benchmark setup
from sklearn.pls import PLSRegression from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('arcene') obj = PLSRegression(**kwargs)
Benchmark statement
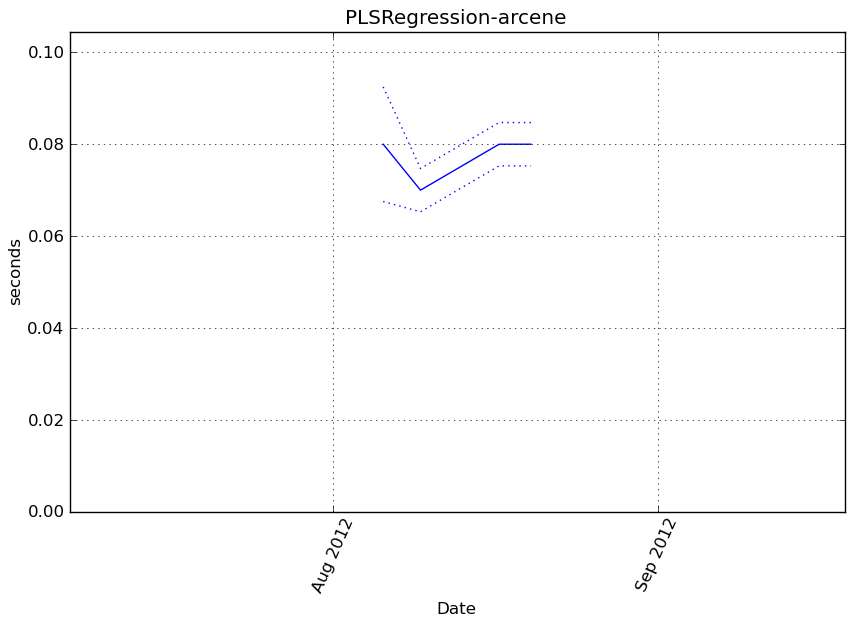
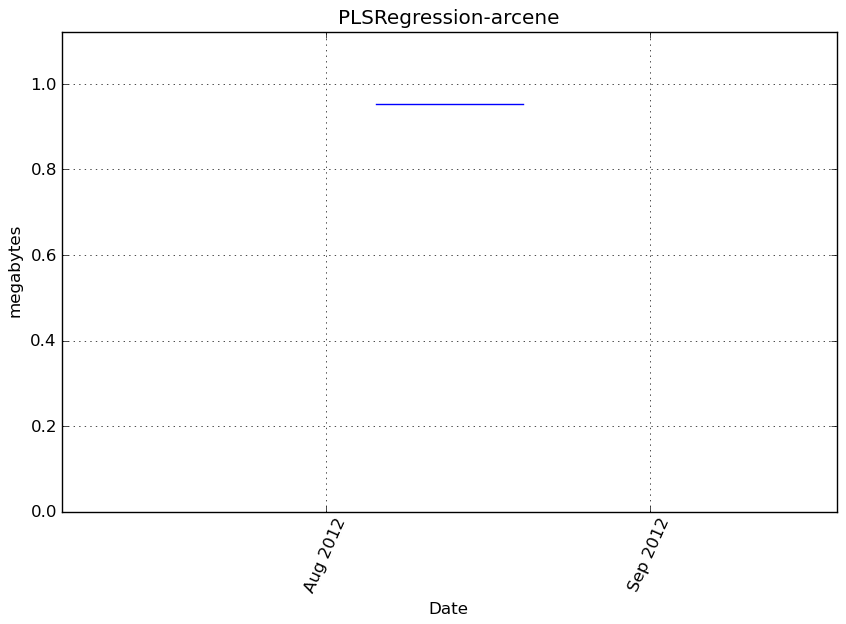
obj.fit(X, y)
Execution time
Memory usage
Additional output
cProfile
113 function calls in 0.095 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.095 0.095 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.095 0.095 <f>:1(<module>)
1 0.008 0.008 0.095 0.095 /tmp/vb_sklearn/sklearn/pls.py:218(fit)
1 0.022 0.022 0.046 0.046 /tmp/vb_sklearn/sklearn/pls.py:75(_center_scale_xy)
41 0.032 0.001 0.032 0.001 {numpy.core._dotblas.dot}
2 0.021 0.010 0.021 0.010 {method 'std' of 'numpy.ndarray' objects}
2 0.002 0.001 0.010 0.005 /tmp/vb_sklearn/sklearn/pls.py:16(_nipals_twoblocks_inner_loop)
2 0.000 0.000 0.006 0.003 /tmp/vb_sklearn/sklearn/utils/validation.py:33(as_float_array)
1 0.006 0.006 0.006 0.006 {method 'copy' of 'numpy.ndarray' objects}
2 0.004 0.002 0.004 0.002 {method 'mean' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:253(inv)
6 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:60(get_lapack_funcs)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py:526(asarray_chkfinite)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:45(find_best_lapack_type)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/getlimits.py:91(__new__)
2 0.000 0.000 0.000 0.000 {method 'any' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
2 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
12 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
2 0.000 0.000 0.000 0.000 {method 'reshape' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:23(cast_to_lapack_prefix)
4 0.000 0.000 0.000 0.000 {getattr}
4 0.000 0.000 0.000 0.000 {isinstance}
1 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
1 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
2 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
2 0.000 0.000 0.000 0.000 {issubclass}
1 0.000 0.000 0.000 0.000 {range}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:449(isfortran)
3 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
1 0.000 0.000 0.000 0.000 {method 'sort' of 'list' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/misc.py:22(_datacopied)
2 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/pls.py
Function: fit at line 218
Total time: 0.084591 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
218 def fit(self, X, Y):
219 # copy since this will contains the residuals (deflated) matrices
220 1 2368 2368.0 2.8 X = as_float_array(X, copy=self.copy)
221 1 47 47.0 0.1 Y = as_float_array(Y, copy=self.copy)
222
223 1 6 6.0 0.0 if X.ndim != 2:
224 raise ValueError('X must be a 2D array')
225 1 4 4.0 0.0 if Y.ndim == 1:
226 1 14 14.0 0.0 Y = Y.reshape((Y.size, 1))
227 1 4 4.0 0.0 if Y.ndim != 2:
228 raise ValueError('Y must be a 1D or a 2D array')
229
230 1 6 6.0 0.0 n = X.shape[0]
231 1 4 4.0 0.0 p = X.shape[1]
232 1 4 4.0 0.0 q = Y.shape[1]
233
234 1 4 4.0 0.0 if n != Y.shape[0]:
235 raise ValueError(
236 'Incompatible shapes: X has %s samples, while Y '
237 'has %s' % (X.shape[0], Y.shape[0]))
238 1 5 5.0 0.0 if self.n_components < 1 or self.n_components > p:
239 raise ValueError('invalid number of components')
240 1 5 5.0 0.0 if self.algorithm not in ("svd", "nipals"):
241 raise ValueError("Got algorithm %s when only 'svd' "
242 "and 'nipals' are known" % self.algorithm)
243 1 3 3.0 0.0 if self.algorithm == "svd" and self.mode == "B":
244 raise ValueError('Incompatible configuration: mode B is not '
245 'implemented with svd algorithm')
246 1 4 4.0 0.0 if not self.deflation_mode in ["canonical", "regression"]:
247 raise ValueError('The deflation mode is unknown')
248 # Scale (in place)
249 X, Y, self.x_mean_, self.y_mean_, self.x_std_, self.y_std_\
250 1 39383 39383.0 46.6 = _center_scale_xy(X, Y, self.scale)
251 # Residuals (deflated) matrices
252 1 5 5.0 0.0 Xk = X
253 1 4 4.0 0.0 Yk = Y
254 # Results matrices
255 1 13 13.0 0.0 self.x_scores_ = np.zeros((n, self.n_components))
256 1 7 7.0 0.0 self.y_scores_ = np.zeros((n, self.n_components))
257 1 35 35.0 0.0 self.x_weights_ = np.zeros((p, self.n_components))
258 1 7 7.0 0.0 self.y_weights_ = np.zeros((q, self.n_components))
259 1 34 34.0 0.0 self.x_loadings_ = np.zeros((p, self.n_components))
260 1 8 8.0 0.0 self.y_loadings_ = np.zeros((q, self.n_components))
261
262 # NIPALS algo: outer loop, over components
263 3 20 6.7 0.0 for k in xrange(self.n_components):
264 #1) weights estimation (inner loop)
265 # -----------------------------------
266 2 10 5.0 0.0 if self.algorithm == "nipals":
267 2 9 4.5 0.0 x_weights, y_weights = _nipals_twoblocks_inner_loop(
268 2 9 4.5 0.0 X=Xk, Y=Yk, mode=self.mode,
269 2 9 4.5 0.0 max_iter=self.max_iter, tol=self.tol,
270 2 10027 5013.5 11.9 norm_y_weights=self.norm_y_weights)
271 elif self.algorithm == "svd":
272 x_weights, y_weights = _svd_cross_product(X=Xk, Y=Yk)
273 # compute scores
274 2 3571 1785.5 4.2 x_scores = np.dot(Xk, x_weights)
275 2 15 7.5 0.0 if self.norm_y_weights:
276 y_ss = 1
277 else:
278 2 40 20.0 0.0 y_ss = np.dot(y_weights.T, y_weights)
279 2 93 46.5 0.1 y_scores = np.dot(Yk, y_weights) / y_ss
280 # test for null variance
281 2 119 59.5 0.1 if np.dot(x_scores.T, x_scores) < np.finfo(np.double).eps:
282 warnings.warn('X scores are null at iteration %s' % k)
283 #2) Deflation (in place)
284 # ----------------------
285 # Possible memory footprint reduction may done here: in order to
286 # avoid the allocation of a data chunk for the rank-one
287 # approximations matrix which is then substracted to Xk, we suggest
288 # to perform a column-wise deflation.
289 #
290 # - regress Xk's on x_score
291 2 5121 2560.5 6.1 x_loadings = np.dot(Xk.T, x_scores) / np.dot(x_scores.T, x_scores)
292 # - substract rank-one approximations to obtain remainder matrix
293 2 21185 10592.5 25.0 Xk -= np.dot(x_scores, x_loadings.T)
294 2 19 9.5 0.0 if self.deflation_mode == "canonical":
295 # - regress Yk's on y_score, then substract rank-one approx.
296 y_loadings = np.dot(Yk.T, y_scores) \
297 / np.dot(y_scores.T, y_scores)
298 Yk -= np.dot(y_scores, y_loadings.T)
299 2 9 4.5 0.0 if self.deflation_mode == "regression":
300 # - regress Yk's on x_score, then substract rank-one approx.
301 2 87 43.5 0.1 y_loadings = np.dot(Yk.T, x_scores) \
302 2 57 28.5 0.1 / np.dot(x_scores.T, x_scores)
303 2 68 34.0 0.1 Yk -= np.dot(x_scores, y_loadings.T)
304 # 3) Store weights, scores and loadings # Notation:
305 2 67 33.5 0.1 self.x_scores_[:, k] = x_scores.ravel() # T
306 2 43 21.5 0.1 self.y_scores_[:, k] = y_scores.ravel() # U
307 2 109 54.5 0.1 self.x_weights_[:, k] = x_weights.ravel() # W
308 2 27 13.5 0.0 self.y_weights_[:, k] = y_weights.ravel() # C
309 2 197 98.5 0.2 self.x_loadings_[:, k] = x_loadings.ravel() # P
310 2 33 16.5 0.0 self.y_loadings_[:, k] = y_loadings.ravel() # Q
311 # Such that: X = TP' + Err and Y = UQ' + Err
312
313 # 4) rotations from input space to transformed space (scores)
314 # T = X W(P'W)^-1 = XW* (W* : p x k matrix)
315 # U = Y C(Q'C)^-1 = YC* (W* : q x k matrix)
316 1 5 5.0 0.0 self.x_rotations_ = np.dot(self.x_weights_,
317 1 1230 1230.0 1.5 linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
318 1 7 7.0 0.0 if Y.shape[1] > 1:
319 self.y_rotations_ = np.dot(self.y_weights_,
320 linalg.inv(np.dot(self.y_loadings_.T, self.y_weights_)))
321 else:
322 1 32 32.0 0.0 self.y_rotations_ = np.ones(1)
323
324 1 5 5.0 0.0 if True or self.deflation_mode == "regression":
325 # Estimate regression coefficient
326 # Regress Y on T
327 # Y = TQ' + Err,
328 # Then express in function of X
329 # Y = X W(P'W)^-1Q' + Err = XB + Err
330 # => B = W*Q' (p x q)
331 1 85 85.0 0.1 self.coefs = np.dot(self.x_rotations_, self.y_loadings_.T)
332 self.coefs = 1. / self.x_std_.reshape((p, 1)) * \
333 1 305 305.0 0.4 self.coefs * self.y_std_
334 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/pls.py
Function: transform at line 336
Total time: 0 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
336 def transform(self, X, Y=None, copy=True):
337 """Apply the dimension reduction learned on the train data.
338
339 Parameters
340 ----------
341 X : array-like of predictors, shape = [n_samples, p]
342 Training vectors, where n_samples in the number of samples and
343 p is the number of predictors.
344
345 Y : array-like of response, shape = [n_samples, q], optional
346 Training vectors, where n_samples in the number of samples and
347 q is the number of response variables.
348
349 copy : boolean
350 Whether to copy X and Y, or perform in-place normalization.
351
352 Returns
353 -------
354 x_scores if Y is not given, (x_scores, y_scores) otherwise.
355 """
356 # Normalize
357 if copy:
358 Xc = (np.asarray(X) - self.x_mean_) / self.x_std_
359 if Y is not None:
360 Yc = (np.asarray(Y) - self.y_mean_) / self.y_std_
361 else:
362 X = np.asarray(X)
363 Xc -= self.x_mean_
364 Xc /= self.x_std_
365 if Y is not None:
366 Y = np.asarray(Y)
367 Yc -= self.y_mean_
368 Yc /= self.y_std_
369 # Apply rotation
370 x_scores = np.dot(Xc, self.x_rotations_)
371 if Y is not None:
372 y_scores = np.dot(Yc, self.y_rotations_)
373 return x_scores, y_scores
374
375 return x_scores
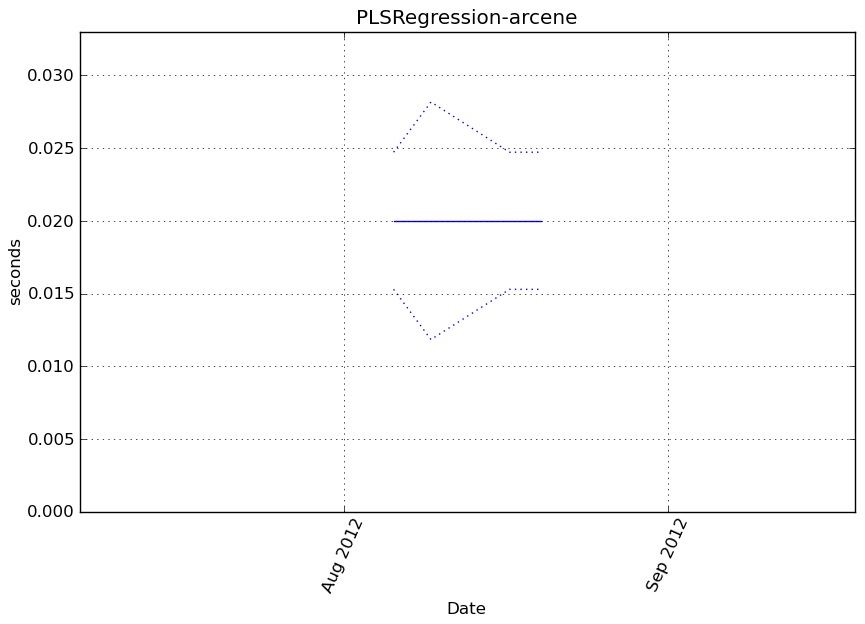
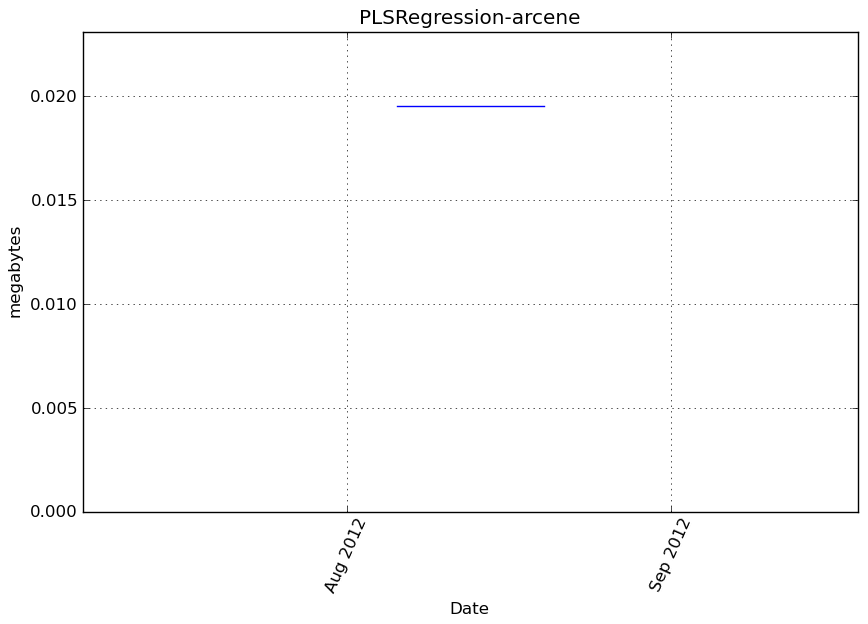
Benchmark statement
obj.transform(X)
Execution time
Memory usage
Additional output
cProfile
7 function calls in 0.029 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.029 0.029 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.029 0.029 <f>:1(<module>)
1 0.022 0.022 0.029 0.029 /tmp/vb_sklearn/sklearn/pls.py:336(transform)
1 0.007 0.007 0.007 0.007 {numpy.core._dotblas.dot}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/pls.py
Function: fit at line 218
Total time: 0.084591 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
218 def fit(self, X, Y):
219 # copy since this will contains the residuals (deflated) matrices
220 1 2368 2368.0 2.8 X = as_float_array(X, copy=self.copy)
221 1 47 47.0 0.1 Y = as_float_array(Y, copy=self.copy)
222
223 1 6 6.0 0.0 if X.ndim != 2:
224 raise ValueError('X must be a 2D array')
225 1 4 4.0 0.0 if Y.ndim == 1:
226 1 14 14.0 0.0 Y = Y.reshape((Y.size, 1))
227 1 4 4.0 0.0 if Y.ndim != 2:
228 raise ValueError('Y must be a 1D or a 2D array')
229
230 1 6 6.0 0.0 n = X.shape[0]
231 1 4 4.0 0.0 p = X.shape[1]
232 1 4 4.0 0.0 q = Y.shape[1]
233
234 1 4 4.0 0.0 if n != Y.shape[0]:
235 raise ValueError(
236 'Incompatible shapes: X has %s samples, while Y '
237 'has %s' % (X.shape[0], Y.shape[0]))
238 1 5 5.0 0.0 if self.n_components < 1 or self.n_components > p:
239 raise ValueError('invalid number of components')
240 1 5 5.0 0.0 if self.algorithm not in ("svd", "nipals"):
241 raise ValueError("Got algorithm %s when only 'svd' "
242 "and 'nipals' are known" % self.algorithm)
243 1 3 3.0 0.0 if self.algorithm == "svd" and self.mode == "B":
244 raise ValueError('Incompatible configuration: mode B is not '
245 'implemented with svd algorithm')
246 1 4 4.0 0.0 if not self.deflation_mode in ["canonical", "regression"]:
247 raise ValueError('The deflation mode is unknown')
248 # Scale (in place)
249 X, Y, self.x_mean_, self.y_mean_, self.x_std_, self.y_std_\
250 1 39383 39383.0 46.6 = _center_scale_xy(X, Y, self.scale)
251 # Residuals (deflated) matrices
252 1 5 5.0 0.0 Xk = X
253 1 4 4.0 0.0 Yk = Y
254 # Results matrices
255 1 13 13.0 0.0 self.x_scores_ = np.zeros((n, self.n_components))
256 1 7 7.0 0.0 self.y_scores_ = np.zeros((n, self.n_components))
257 1 35 35.0 0.0 self.x_weights_ = np.zeros((p, self.n_components))
258 1 7 7.0 0.0 self.y_weights_ = np.zeros((q, self.n_components))
259 1 34 34.0 0.0 self.x_loadings_ = np.zeros((p, self.n_components))
260 1 8 8.0 0.0 self.y_loadings_ = np.zeros((q, self.n_components))
261
262 # NIPALS algo: outer loop, over components
263 3 20 6.7 0.0 for k in xrange(self.n_components):
264 #1) weights estimation (inner loop)
265 # -----------------------------------
266 2 10 5.0 0.0 if self.algorithm == "nipals":
267 2 9 4.5 0.0 x_weights, y_weights = _nipals_twoblocks_inner_loop(
268 2 9 4.5 0.0 X=Xk, Y=Yk, mode=self.mode,
269 2 9 4.5 0.0 max_iter=self.max_iter, tol=self.tol,
270 2 10027 5013.5 11.9 norm_y_weights=self.norm_y_weights)
271 elif self.algorithm == "svd":
272 x_weights, y_weights = _svd_cross_product(X=Xk, Y=Yk)
273 # compute scores
274 2 3571 1785.5 4.2 x_scores = np.dot(Xk, x_weights)
275 2 15 7.5 0.0 if self.norm_y_weights:
276 y_ss = 1
277 else:
278 2 40 20.0 0.0 y_ss = np.dot(y_weights.T, y_weights)
279 2 93 46.5 0.1 y_scores = np.dot(Yk, y_weights) / y_ss
280 # test for null variance
281 2 119 59.5 0.1 if np.dot(x_scores.T, x_scores) < np.finfo(np.double).eps:
282 warnings.warn('X scores are null at iteration %s' % k)
283 #2) Deflation (in place)
284 # ----------------------
285 # Possible memory footprint reduction may done here: in order to
286 # avoid the allocation of a data chunk for the rank-one
287 # approximations matrix which is then substracted to Xk, we suggest
288 # to perform a column-wise deflation.
289 #
290 # - regress Xk's on x_score
291 2 5121 2560.5 6.1 x_loadings = np.dot(Xk.T, x_scores) / np.dot(x_scores.T, x_scores)
292 # - substract rank-one approximations to obtain remainder matrix
293 2 21185 10592.5 25.0 Xk -= np.dot(x_scores, x_loadings.T)
294 2 19 9.5 0.0 if self.deflation_mode == "canonical":
295 # - regress Yk's on y_score, then substract rank-one approx.
296 y_loadings = np.dot(Yk.T, y_scores) \
297 / np.dot(y_scores.T, y_scores)
298 Yk -= np.dot(y_scores, y_loadings.T)
299 2 9 4.5 0.0 if self.deflation_mode == "regression":
300 # - regress Yk's on x_score, then substract rank-one approx.
301 2 87 43.5 0.1 y_loadings = np.dot(Yk.T, x_scores) \
302 2 57 28.5 0.1 / np.dot(x_scores.T, x_scores)
303 2 68 34.0 0.1 Yk -= np.dot(x_scores, y_loadings.T)
304 # 3) Store weights, scores and loadings # Notation:
305 2 67 33.5 0.1 self.x_scores_[:, k] = x_scores.ravel() # T
306 2 43 21.5 0.1 self.y_scores_[:, k] = y_scores.ravel() # U
307 2 109 54.5 0.1 self.x_weights_[:, k] = x_weights.ravel() # W
308 2 27 13.5 0.0 self.y_weights_[:, k] = y_weights.ravel() # C
309 2 197 98.5 0.2 self.x_loadings_[:, k] = x_loadings.ravel() # P
310 2 33 16.5 0.0 self.y_loadings_[:, k] = y_loadings.ravel() # Q
311 # Such that: X = TP' + Err and Y = UQ' + Err
312
313 # 4) rotations from input space to transformed space (scores)
314 # T = X W(P'W)^-1 = XW* (W* : p x k matrix)
315 # U = Y C(Q'C)^-1 = YC* (W* : q x k matrix)
316 1 5 5.0 0.0 self.x_rotations_ = np.dot(self.x_weights_,
317 1 1230 1230.0 1.5 linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
318 1 7 7.0 0.0 if Y.shape[1] > 1:
319 self.y_rotations_ = np.dot(self.y_weights_,
320 linalg.inv(np.dot(self.y_loadings_.T, self.y_weights_)))
321 else:
322 1 32 32.0 0.0 self.y_rotations_ = np.ones(1)
323
324 1 5 5.0 0.0 if True or self.deflation_mode == "regression":
325 # Estimate regression coefficient
326 # Regress Y on T
327 # Y = TQ' + Err,
328 # Then express in function of X
329 # Y = X W(P'W)^-1Q' + Err = XB + Err
330 # => B = W*Q' (p x q)
331 1 85 85.0 0.1 self.coefs = np.dot(self.x_rotations_, self.y_loadings_.T)
332 self.coefs = 1. / self.x_std_.reshape((p, 1)) * \
333 1 305 305.0 0.4 self.coefs * self.y_std_
334 1 4 4.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/pls.py
Function: transform at line 336
Total time: 0.028822 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
336 def transform(self, X, Y=None, copy=True):
337 """Apply the dimension reduction learned on the train data.
338
339 Parameters
340 ----------
341 X : array-like of predictors, shape = [n_samples, p]
342 Training vectors, where n_samples in the number of samples and
343 p is the number of predictors.
344
345 Y : array-like of response, shape = [n_samples, q], optional
346 Training vectors, where n_samples in the number of samples and
347 q is the number of response variables.
348
349 copy : boolean
350 Whether to copy X and Y, or perform in-place normalization.
351
352 Returns
353 -------
354 x_scores if Y is not given, (x_scores, y_scores) otherwise.
355 """
356 # Normalize
357 1 3 3.0 0.0 if copy:
358 1 22042 22042.0 76.5 Xc = (np.asarray(X) - self.x_mean_) / self.x_std_
359 1 4 4.0 0.0 if Y is not None:
360 Yc = (np.asarray(Y) - self.y_mean_) / self.y_std_
361 else:
362 X = np.asarray(X)
363 Xc -= self.x_mean_
364 Xc /= self.x_std_
365 if Y is not None:
366 Y = np.asarray(Y)
367 Yc -= self.y_mean_
368 Yc /= self.y_std_
369 # Apply rotation
370 1 6767 6767.0 23.5 x_scores = np.dot(Xc, self.x_rotations_)
371 1 4 4.0 0.0 if Y is not None:
372 y_scores = np.dot(Yc, self.y_rotations_)
373 return x_scores, y_scores
374
375 1 2 2.0 0.0 return x_scores
PLSRegression-madelon¶
Benchmark setup
from sklearn.pls import PLSRegression from deps import load_data kwargs = {} X, y, X_t, y_t = load_data('madelon') obj = PLSRegression(**kwargs)
Benchmark statement
obj.fit(X, y)
Execution time
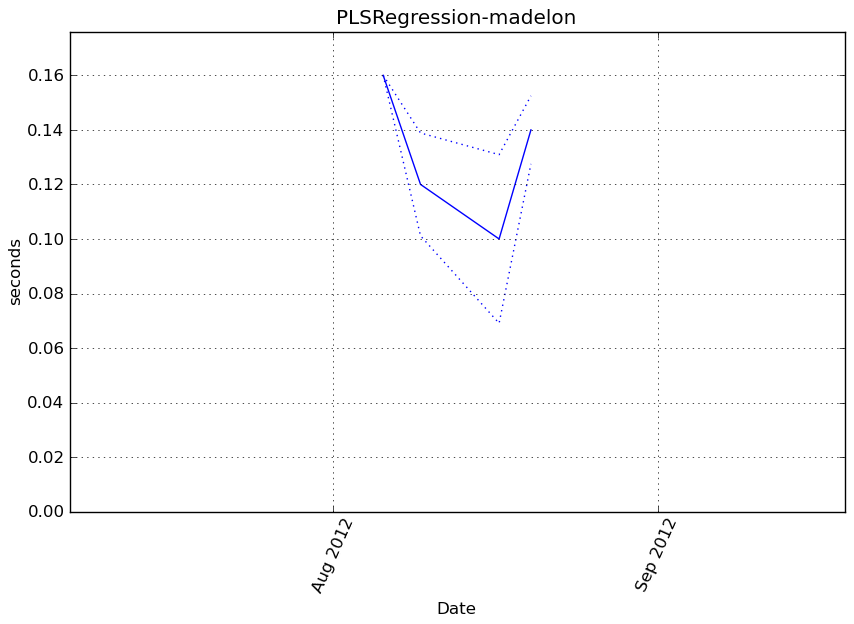
Memory usage
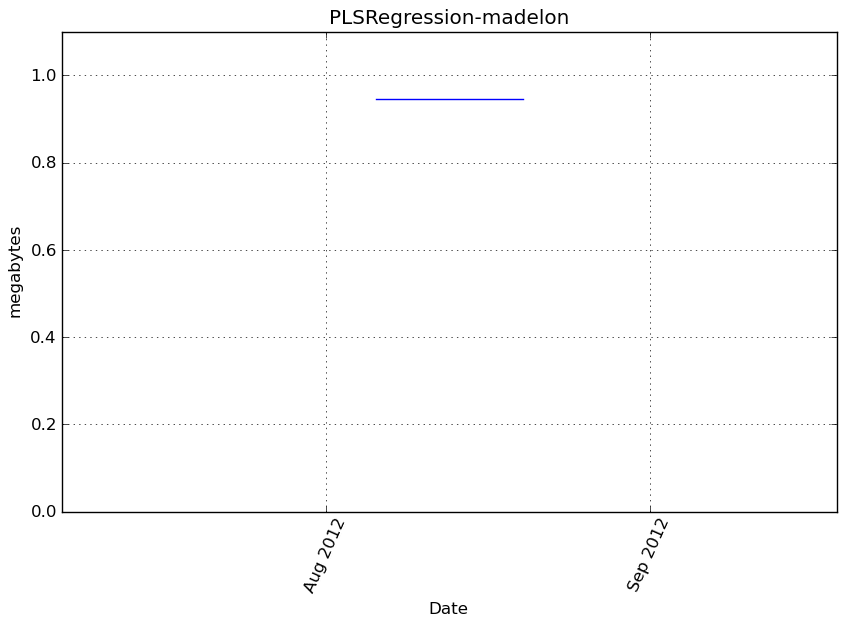
Additional output
cProfile
113 function calls in 0.104 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.104 0.104 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.104 0.104 <f>:1(<module>)
1 0.006 0.006 0.104 0.104 /tmp/vb_sklearn/sklearn/pls.py:218(fit)
1 0.013 0.013 0.073 0.073 /tmp/vb_sklearn/sklearn/pls.py:75(_center_scale_xy)
2 0.043 0.022 0.043 0.022 {method 'std' of 'numpy.ndarray' objects}
41 0.020 0.000 0.020 0.000 {numpy.core._dotblas.dot}
2 0.017 0.009 0.017 0.009 {method 'mean' of 'numpy.ndarray' objects}
2 0.000 0.000 0.006 0.003 /tmp/vb_sklearn/sklearn/pls.py:16(_nipals_twoblocks_inner_loop)
2 0.000 0.000 0.004 0.002 /tmp/vb_sklearn/sklearn/utils/validation.py:33(as_float_array)
1 0.004 0.004 0.004 0.004 {method 'copy' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/basic.py:253(inv)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:60(get_lapack_funcs)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/lib/function_base.py:526(asarray_chkfinite)
2 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/getlimits.py:91(__new__)
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:45(find_best_lapack_type)
6 0.000 0.000 0.000 0.000 {numpy.core.multiarray.zeros}
2 0.000 0.000 0.000 0.000 {method 'any' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:1791(ones)
2 0.000 0.000 0.000 0.000 {method 'get' of 'dict' objects}
12 0.000 0.000 0.000 0.000 {method 'ravel' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 {method 'astype' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 {isinstance}
2 0.000 0.000 0.000 0.000 {method 'reshape' of 'numpy.ndarray' objects}
4 0.000 0.000 0.000 0.000 {getattr}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/lapack.py:23(cast_to_lapack_prefix)
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.empty}
1 0.000 0.000 0.000 0.000 {method 'fill' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
2 0.000 0.000 0.000 0.000 {method 'split' of 'str' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:449(isfortran)
2 0.000 0.000 0.000 0.000 {issubclass}
1 0.000 0.000 0.000 0.000 {range}
3 0.000 0.000 0.000 0.000 {method 'append' of 'list' objects}
1 0.000 0.000 0.000 0.000 {method 'sort' of 'list' objects}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/scipy/linalg/misc.py:22(_datacopied)
2 0.000 0.000 0.000 0.000 {len}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/pls.py
Function: fit at line 218
Total time: 0.098747 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
218 def fit(self, X, Y):
219 # copy since this will contains the residuals (deflated) matrices
220 1 1753 1753.0 1.8 X = as_float_array(X, copy=self.copy)
221 1 35 35.0 0.0 Y = as_float_array(Y, copy=self.copy)
222
223 1 4 4.0 0.0 if X.ndim != 2:
224 raise ValueError('X must be a 2D array')
225 1 2 2.0 0.0 if Y.ndim == 1:
226 1 9 9.0 0.0 Y = Y.reshape((Y.size, 1))
227 1 2 2.0 0.0 if Y.ndim != 2:
228 raise ValueError('Y must be a 1D or a 2D array')
229
230 1 4 4.0 0.0 n = X.shape[0]
231 1 3 3.0 0.0 p = X.shape[1]
232 1 3 3.0 0.0 q = Y.shape[1]
233
234 1 3 3.0 0.0 if n != Y.shape[0]:
235 raise ValueError(
236 'Incompatible shapes: X has %s samples, while Y '
237 'has %s' % (X.shape[0], Y.shape[0]))
238 1 3 3.0 0.0 if self.n_components < 1 or self.n_components > p:
239 raise ValueError('invalid number of components')
240 1 3 3.0 0.0 if self.algorithm not in ("svd", "nipals"):
241 raise ValueError("Got algorithm %s when only 'svd' "
242 "and 'nipals' are known" % self.algorithm)
243 1 2 2.0 0.0 if self.algorithm == "svd" and self.mode == "B":
244 raise ValueError('Incompatible configuration: mode B is not '
245 'implemented with svd algorithm')
246 1 2 2.0 0.0 if not self.deflation_mode in ["canonical", "regression"]:
247 raise ValueError('The deflation mode is unknown')
248 # Scale (in place)
249 X, Y, self.x_mean_, self.y_mean_, self.x_std_, self.y_std_\
250 1 69556 69556.0 70.4 = _center_scale_xy(X, Y, self.scale)
251 # Residuals (deflated) matrices
252 1 3 3.0 0.0 Xk = X
253 1 2 2.0 0.0 Yk = Y
254 # Results matrices
255 1 14 14.0 0.0 self.x_scores_ = np.zeros((n, self.n_components))
256 1 9 9.0 0.0 self.y_scores_ = np.zeros((n, self.n_components))
257 1 5 5.0 0.0 self.x_weights_ = np.zeros((p, self.n_components))
258 1 4 4.0 0.0 self.y_weights_ = np.zeros((q, self.n_components))
259 1 5 5.0 0.0 self.x_loadings_ = np.zeros((p, self.n_components))
260 1 4 4.0 0.0 self.y_loadings_ = np.zeros((q, self.n_components))
261
262 # NIPALS algo: outer loop, over components
263 3 13 4.3 0.0 for k in xrange(self.n_components):
264 #1) weights estimation (inner loop)
265 # -----------------------------------
266 2 6 3.0 0.0 if self.algorithm == "nipals":
267 2 5 2.5 0.0 x_weights, y_weights = _nipals_twoblocks_inner_loop(
268 2 5 2.5 0.0 X=Xk, Y=Yk, mode=self.mode,
269 2 4 2.0 0.0 max_iter=self.max_iter, tol=self.tol,
270 2 6191 3095.5 6.3 norm_y_weights=self.norm_y_weights)
271 elif self.algorithm == "svd":
272 x_weights, y_weights = _svd_cross_product(X=Xk, Y=Yk)
273 # compute scores
274 2 2154 1077.0 2.2 x_scores = np.dot(Xk, x_weights)
275 2 9 4.5 0.0 if self.norm_y_weights:
276 y_ss = 1
277 else:
278 2 26 13.0 0.0 y_ss = np.dot(y_weights.T, y_weights)
279 2 115 57.5 0.1 y_scores = np.dot(Yk, y_weights) / y_ss
280 # test for null variance
281 2 83 41.5 0.1 if np.dot(x_scores.T, x_scores) < np.finfo(np.double).eps:
282 warnings.warn('X scores are null at iteration %s' % k)
283 #2) Deflation (in place)
284 # ----------------------
285 # Possible memory footprint reduction may done here: in order to
286 # avoid the allocation of a data chunk for the rank-one
287 # approximations matrix which is then substracted to Xk, we suggest
288 # to perform a column-wise deflation.
289 #
290 # - regress Xk's on x_score
291 2 2822 1411.0 2.9 x_loadings = np.dot(Xk.T, x_scores) / np.dot(x_scores.T, x_scores)
292 # - substract rank-one approximations to obtain remainder matrix
293 2 15031 7515.5 15.2 Xk -= np.dot(x_scores, x_loadings.T)
294 2 13 6.5 0.0 if self.deflation_mode == "canonical":
295 # - regress Yk's on y_score, then substract rank-one approx.
296 y_loadings = np.dot(Yk.T, y_scores) \
297 / np.dot(y_scores.T, y_scores)
298 Yk -= np.dot(y_scores, y_loadings.T)
299 2 5 2.5 0.0 if self.deflation_mode == "regression":
300 # - regress Yk's on x_score, then substract rank-one approx.
301 2 80 40.0 0.1 y_loadings = np.dot(Yk.T, x_scores) \
302 2 44 22.0 0.0 / np.dot(x_scores.T, x_scores)
303 2 76 38.0 0.1 Yk -= np.dot(x_scores, y_loadings.T)
304 # 3) Store weights, scores and loadings # Notation:
305 2 56 28.0 0.1 self.x_scores_[:, k] = x_scores.ravel() # T
306 2 30 15.0 0.0 self.y_scores_[:, k] = y_scores.ravel() # U
307 2 27 13.5 0.0 self.x_weights_[:, k] = x_weights.ravel() # W
308 2 15 7.5 0.0 self.y_weights_[:, k] = y_weights.ravel() # C
309 2 21 10.5 0.0 self.x_loadings_[:, k] = x_loadings.ravel() # P
310 2 12 6.0 0.0 self.y_loadings_[:, k] = y_loadings.ravel() # Q
311 # Such that: X = TP' + Err and Y = UQ' + Err
312
313 # 4) rotations from input space to transformed space (scores)
314 # T = X W(P'W)^-1 = XW* (W* : p x k matrix)
315 # U = Y C(Q'C)^-1 = YC* (W* : q x k matrix)
316 1 2 2.0 0.0 self.x_rotations_ = np.dot(self.x_weights_,
317 1 386 386.0 0.4 linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
318 1 4 4.0 0.0 if Y.shape[1] > 1:
319 self.y_rotations_ = np.dot(self.y_weights_,
320 linalg.inv(np.dot(self.y_loadings_.T, self.y_weights_)))
321 else:
322 1 21 21.0 0.0 self.y_rotations_ = np.ones(1)
323
324 1 2 2.0 0.0 if True or self.deflation_mode == "regression":
325 # Estimate regression coefficient
326 # Regress Y on T
327 # Y = TQ' + Err,
328 # Then express in function of X
329 # Y = X W(P'W)^-1Q' + Err = XB + Err
330 # => B = W*Q' (p x q)
331 1 14 14.0 0.0 self.coefs = np.dot(self.x_rotations_, self.y_loadings_.T)
332 self.coefs = 1. / self.x_std_.reshape((p, 1)) * \
333 1 42 42.0 0.0 self.coefs * self.y_std_
334 1 3 3.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/pls.py
Function: transform at line 336
Total time: 0 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
336 def transform(self, X, Y=None, copy=True):
337 """Apply the dimension reduction learned on the train data.
338
339 Parameters
340 ----------
341 X : array-like of predictors, shape = [n_samples, p]
342 Training vectors, where n_samples in the number of samples and
343 p is the number of predictors.
344
345 Y : array-like of response, shape = [n_samples, q], optional
346 Training vectors, where n_samples in the number of samples and
347 q is the number of response variables.
348
349 copy : boolean
350 Whether to copy X and Y, or perform in-place normalization.
351
352 Returns
353 -------
354 x_scores if Y is not given, (x_scores, y_scores) otherwise.
355 """
356 # Normalize
357 if copy:
358 Xc = (np.asarray(X) - self.x_mean_) / self.x_std_
359 if Y is not None:
360 Yc = (np.asarray(Y) - self.y_mean_) / self.y_std_
361 else:
362 X = np.asarray(X)
363 Xc -= self.x_mean_
364 Xc /= self.x_std_
365 if Y is not None:
366 Y = np.asarray(Y)
367 Yc -= self.y_mean_
368 Yc /= self.y_std_
369 # Apply rotation
370 x_scores = np.dot(Xc, self.x_rotations_)
371 if Y is not None:
372 y_scores = np.dot(Yc, self.y_rotations_)
373 return x_scores, y_scores
374
375 return x_scores
Benchmark statement
obj.transform(X)
Execution time
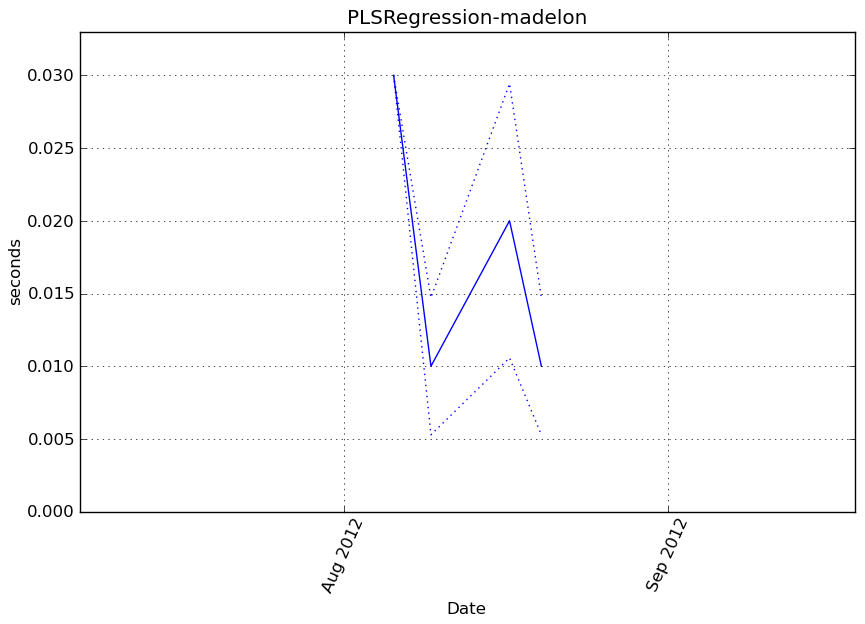
Memory usage
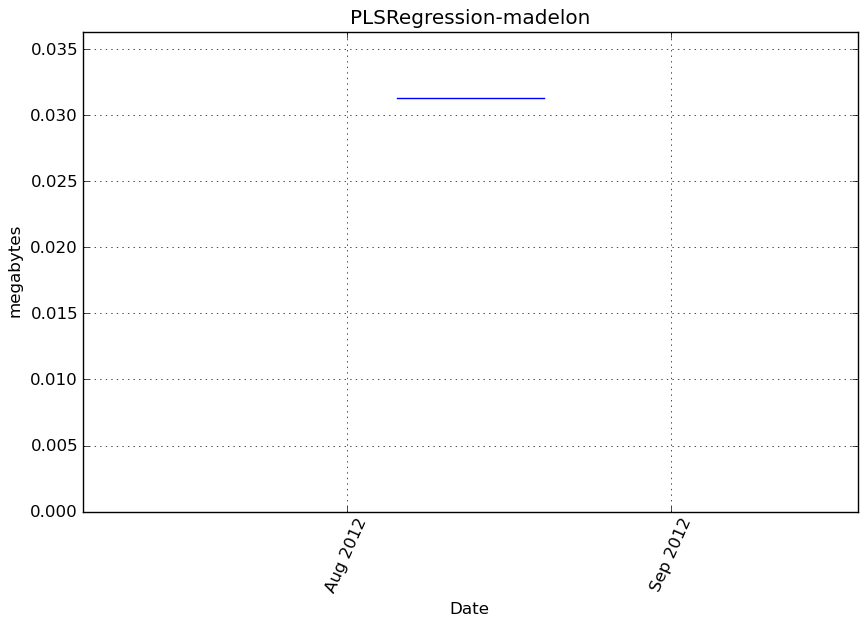
Additional output
cProfile
7 function calls in 0.018 seconds
Ordered by: cumulative time
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.018 0.018 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/vbench/benchmark.py:286(f)
1 0.000 0.000 0.018 0.018 <f>:1(<module>)
1 0.014 0.014 0.018 0.018 /tmp/vb_sklearn/sklearn/pls.py:336(transform)
1 0.004 0.004 0.004 0.004 {numpy.core._dotblas.dot}
1 0.000 0.000 0.000 0.000 /home/slave/virtualenvs/cpython-2.7.2/lib/python2.7/site-packages/numpy/core/numeric.py:167(asarray)
1 0.000 0.000 0.000 0.000 {numpy.core.multiarray.array}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
LineProfiler
Timer unit: 1e-06 s
File: /tmp/vb_sklearn/sklearn/pls.py
Function: fit at line 218
Total time: 0.098747 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
218 def fit(self, X, Y):
219 # copy since this will contains the residuals (deflated) matrices
220 1 1753 1753.0 1.8 X = as_float_array(X, copy=self.copy)
221 1 35 35.0 0.0 Y = as_float_array(Y, copy=self.copy)
222
223 1 4 4.0 0.0 if X.ndim != 2:
224 raise ValueError('X must be a 2D array')
225 1 2 2.0 0.0 if Y.ndim == 1:
226 1 9 9.0 0.0 Y = Y.reshape((Y.size, 1))
227 1 2 2.0 0.0 if Y.ndim != 2:
228 raise ValueError('Y must be a 1D or a 2D array')
229
230 1 4 4.0 0.0 n = X.shape[0]
231 1 3 3.0 0.0 p = X.shape[1]
232 1 3 3.0 0.0 q = Y.shape[1]
233
234 1 3 3.0 0.0 if n != Y.shape[0]:
235 raise ValueError(
236 'Incompatible shapes: X has %s samples, while Y '
237 'has %s' % (X.shape[0], Y.shape[0]))
238 1 3 3.0 0.0 if self.n_components < 1 or self.n_components > p:
239 raise ValueError('invalid number of components')
240 1 3 3.0 0.0 if self.algorithm not in ("svd", "nipals"):
241 raise ValueError("Got algorithm %s when only 'svd' "
242 "and 'nipals' are known" % self.algorithm)
243 1 2 2.0 0.0 if self.algorithm == "svd" and self.mode == "B":
244 raise ValueError('Incompatible configuration: mode B is not '
245 'implemented with svd algorithm')
246 1 2 2.0 0.0 if not self.deflation_mode in ["canonical", "regression"]:
247 raise ValueError('The deflation mode is unknown')
248 # Scale (in place)
249 X, Y, self.x_mean_, self.y_mean_, self.x_std_, self.y_std_\
250 1 69556 69556.0 70.4 = _center_scale_xy(X, Y, self.scale)
251 # Residuals (deflated) matrices
252 1 3 3.0 0.0 Xk = X
253 1 2 2.0 0.0 Yk = Y
254 # Results matrices
255 1 14 14.0 0.0 self.x_scores_ = np.zeros((n, self.n_components))
256 1 9 9.0 0.0 self.y_scores_ = np.zeros((n, self.n_components))
257 1 5 5.0 0.0 self.x_weights_ = np.zeros((p, self.n_components))
258 1 4 4.0 0.0 self.y_weights_ = np.zeros((q, self.n_components))
259 1 5 5.0 0.0 self.x_loadings_ = np.zeros((p, self.n_components))
260 1 4 4.0 0.0 self.y_loadings_ = np.zeros((q, self.n_components))
261
262 # NIPALS algo: outer loop, over components
263 3 13 4.3 0.0 for k in xrange(self.n_components):
264 #1) weights estimation (inner loop)
265 # -----------------------------------
266 2 6 3.0 0.0 if self.algorithm == "nipals":
267 2 5 2.5 0.0 x_weights, y_weights = _nipals_twoblocks_inner_loop(
268 2 5 2.5 0.0 X=Xk, Y=Yk, mode=self.mode,
269 2 4 2.0 0.0 max_iter=self.max_iter, tol=self.tol,
270 2 6191 3095.5 6.3 norm_y_weights=self.norm_y_weights)
271 elif self.algorithm == "svd":
272 x_weights, y_weights = _svd_cross_product(X=Xk, Y=Yk)
273 # compute scores
274 2 2154 1077.0 2.2 x_scores = np.dot(Xk, x_weights)
275 2 9 4.5 0.0 if self.norm_y_weights:
276 y_ss = 1
277 else:
278 2 26 13.0 0.0 y_ss = np.dot(y_weights.T, y_weights)
279 2 115 57.5 0.1 y_scores = np.dot(Yk, y_weights) / y_ss
280 # test for null variance
281 2 83 41.5 0.1 if np.dot(x_scores.T, x_scores) < np.finfo(np.double).eps:
282 warnings.warn('X scores are null at iteration %s' % k)
283 #2) Deflation (in place)
284 # ----------------------
285 # Possible memory footprint reduction may done here: in order to
286 # avoid the allocation of a data chunk for the rank-one
287 # approximations matrix which is then substracted to Xk, we suggest
288 # to perform a column-wise deflation.
289 #
290 # - regress Xk's on x_score
291 2 2822 1411.0 2.9 x_loadings = np.dot(Xk.T, x_scores) / np.dot(x_scores.T, x_scores)
292 # - substract rank-one approximations to obtain remainder matrix
293 2 15031 7515.5 15.2 Xk -= np.dot(x_scores, x_loadings.T)
294 2 13 6.5 0.0 if self.deflation_mode == "canonical":
295 # - regress Yk's on y_score, then substract rank-one approx.
296 y_loadings = np.dot(Yk.T, y_scores) \
297 / np.dot(y_scores.T, y_scores)
298 Yk -= np.dot(y_scores, y_loadings.T)
299 2 5 2.5 0.0 if self.deflation_mode == "regression":
300 # - regress Yk's on x_score, then substract rank-one approx.
301 2 80 40.0 0.1 y_loadings = np.dot(Yk.T, x_scores) \
302 2 44 22.0 0.0 / np.dot(x_scores.T, x_scores)
303 2 76 38.0 0.1 Yk -= np.dot(x_scores, y_loadings.T)
304 # 3) Store weights, scores and loadings # Notation:
305 2 56 28.0 0.1 self.x_scores_[:, k] = x_scores.ravel() # T
306 2 30 15.0 0.0 self.y_scores_[:, k] = y_scores.ravel() # U
307 2 27 13.5 0.0 self.x_weights_[:, k] = x_weights.ravel() # W
308 2 15 7.5 0.0 self.y_weights_[:, k] = y_weights.ravel() # C
309 2 21 10.5 0.0 self.x_loadings_[:, k] = x_loadings.ravel() # P
310 2 12 6.0 0.0 self.y_loadings_[:, k] = y_loadings.ravel() # Q
311 # Such that: X = TP' + Err and Y = UQ' + Err
312
313 # 4) rotations from input space to transformed space (scores)
314 # T = X W(P'W)^-1 = XW* (W* : p x k matrix)
315 # U = Y C(Q'C)^-1 = YC* (W* : q x k matrix)
316 1 2 2.0 0.0 self.x_rotations_ = np.dot(self.x_weights_,
317 1 386 386.0 0.4 linalg.inv(np.dot(self.x_loadings_.T, self.x_weights_)))
318 1 4 4.0 0.0 if Y.shape[1] > 1:
319 self.y_rotations_ = np.dot(self.y_weights_,
320 linalg.inv(np.dot(self.y_loadings_.T, self.y_weights_)))
321 else:
322 1 21 21.0 0.0 self.y_rotations_ = np.ones(1)
323
324 1 2 2.0 0.0 if True or self.deflation_mode == "regression":
325 # Estimate regression coefficient
326 # Regress Y on T
327 # Y = TQ' + Err,
328 # Then express in function of X
329 # Y = X W(P'W)^-1Q' + Err = XB + Err
330 # => B = W*Q' (p x q)
331 1 14 14.0 0.0 self.coefs = np.dot(self.x_rotations_, self.y_loadings_.T)
332 self.coefs = 1. / self.x_std_.reshape((p, 1)) * \
333 1 42 42.0 0.0 self.coefs * self.y_std_
334 1 3 3.0 0.0 return self
File: /tmp/vb_sklearn/sklearn/pls.py
Function: transform at line 336
Total time: 0.017922 s
Line # Hits Time Per Hit % Time Line Contents
==============================================================
336 def transform(self, X, Y=None, copy=True):
337 """Apply the dimension reduction learned on the train data.
338
339 Parameters
340 ----------
341 X : array-like of predictors, shape = [n_samples, p]
342 Training vectors, where n_samples in the number of samples and
343 p is the number of predictors.
344
345 Y : array-like of response, shape = [n_samples, q], optional
346 Training vectors, where n_samples in the number of samples and
347 q is the number of response variables.
348
349 copy : boolean
350 Whether to copy X and Y, or perform in-place normalization.
351
352 Returns
353 -------
354 x_scores if Y is not given, (x_scores, y_scores) otherwise.
355 """
356 # Normalize
357 1 2 2.0 0.0 if copy:
358 1 13838 13838.0 77.2 Xc = (np.asarray(X) - self.x_mean_) / self.x_std_
359 1 4 4.0 0.0 if Y is not None:
360 Yc = (np.asarray(Y) - self.y_mean_) / self.y_std_
361 else:
362 X = np.asarray(X)
363 Xc -= self.x_mean_
364 Xc /= self.x_std_
365 if Y is not None:
366 Y = np.asarray(Y)
367 Yc -= self.y_mean_
368 Yc /= self.y_std_
369 # Apply rotation
370 1 4075 4075.0 22.7 x_scores = np.dot(Xc, self.x_rotations_)
371 1 2 2.0 0.0 if Y is not None:
372 y_scores = np.dot(Yc, self.y_rotations_)
373 return x_scores, y_scores
374
375 1 1 1.0 0.0 return x_scores
